Handling_missingval_using_knn_02
title: “Handing Missing Categorical Value using Hamming distance” excerpt: “using sckit-learn or handmade Function Test”
categories:
- Useful_Fuction tags:
- KNN
- OrdinalEncoder
- Categorical Missing Value
last_modified_at: 2020-05-23T08:06:00-05:00 —
지난 포스팅은 scikit-learn 0.23.1 에서 출시한, KNNimputer 를 사용했다는 점에서 흥미롭긴 하지만, 원래 목적이었던, categorical 변수에 대해서는 약간의 아쉬움이 남았다.
이번에는 categorical 변수에 대해서, distance 개념으로, 즉, 다른 방법으로 시도했던 자료를 찾아서 포스팅한다.
Reference
Missing Value 의 3가지 type
1. MCAR (missing completely at random): 특정 변수의 결측치가 완전히 무작위적으로 발생할 경우
MCAR when the probability of missing data on a variable X is unrelated to other measured variables and to the values of X itself.
좀 더 정확히는, 변수 x의 결측이 발생하는 확률이 x의 값 자체나 다른 변수들과 관련이 없을 때
2.MAR (missing at random): 특정 변수의 결측의 여부가 자료 내의 다른 변수와 관련이 있는 경우
예를들어 학업성취 점수의 결측 여부가 소득수준과 관련이 있을 때 (즉, 소득수준이 낮은 아이들이 학업성취점수에 응답하지 않음) 예를들어, 남성들이 우울증에 대한 설문에 답변을 하지 않는 경우가 많은데, 이런 결측값들은 실제 우울증 정도와는 무관하다.
3.MNAR (missing not at random, non ignorable): 결측여부가 해당변수의 값에 의해 결정
예를들어 학업성취가 낮은 아이들이 학업성취에 응답하지 않음 예를들어 우울증정도에 따라서, 진짜로 설문조사에 응하지 않을 경우.
대부분의 분석들, 결측자료를 처리하는 방법들(SEM에서 FIML, EM, multiple imputation)은 MCAR, MAR을 가정하고 있음
Distance of Categorical Variable
숫자형으로 labeling 하지 않는다면, 카테고리컬 변수에 distance 개념을 적용하는 방법은 빈도 및 유사성과 관련이 있다.
여기서는 Hamming distance 과 Weighted Hamming distance 를 다룬다.
Hamming distance
- 모든 범주형 속성을 취하며, 두 점(로우) 사이의 값이 같지 않을 경우 각 범주형 속성을 1로 계산한다. 해밍 거리는 그 값이 다른 속성의 수입니다.
Weighted Hamming distance
- 값이 다르면 1을 반환하지만, 일치하면 속성의 값의 빈도를 반환하여 해당 범주값이 더 빈번할 때(자주 나오는 범주값일 경우)거리를 증가시킨다.
- 둘 이상의 속성이 범주형일 경우 조화평균(harmonic)이 적용된다.
- 결과는 0과 1 사이에 유지되지만 평균 값은 산술 평균에 비해 낮은 값으로 이동한다.
import numpy as np
import pandas as pd
from collections import defaultdict
from scipy.stats import hmean
from scipy.spatial.distance import cdist
from scipy import stats
import numbers
import warnings
warnings.filterwarnings("ignore")
train = pd.read_csv('../dataset/titanic_train.csv')
test = pd.read_csv('../dataset/titanic_test.csv')
# train = pd.read_csv('../ML_Area/data_source/titanic_train.csv')
# test = pd.read_csv('../ML_Area/data_source/titanic_test.csv')
# ML_Area\data_source
import matplotlib.pyplot as plt
%matplotlib inline
import seaborn as sns
sns.set() # setting seaborn default for plots
pd.concat([train.isnull().sum(),train.dtypes],axis=1,names=[['null','dtype']])
| 0 | 1 | |
|---|---|---|
| PassengerId | 0 | int64 |
| Survived | 0 | int64 |
| Pclass | 0 | int64 |
| Name | 0 | object |
| Sex | 0 | object |
| Age | 177 | float64 |
| SibSp | 0 | int64 |
| Parch | 0 | int64 |
| Ticket | 0 | object |
| Fare | 0 | float64 |
| Cabin | 687 | object |
| Embarked | 2 | object |
범주형 변수값들의 거리를 구해보자
not_cat_df = train[[col for col in train.columns if col not in ['Cabin','Embarked']]].copy() ## Cabin 제외한 df
int_col = train.columns[train.dtypes != 'object'].to_list()
cat_col = train.columns[train.dtypes == 'object'].to_list()
train.loc[train["Embarked"].isnull(),:]
| PassengerId | Survived | Pclass | Name | Sex | Age | SibSp | Parch | Ticket | Fare | Cabin | Embarked | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 61 | 62 | 1 | 1 | Icard, Miss. Amelie | female | 38.0 | 0 | 0 | 113572 | 80.0 | B28 | NaN |
| 829 | 830 | 1 | 1 | Stone, Mrs. George Nelson (Martha Evelyn) | female | 62.0 | 0 | 0 | 113572 | 80.0 | B28 | NaN |
## 나중에 확인할 데이터 영역
train.iloc[60:65]
| PassengerId | Survived | Pclass | Name | Sex | Age | SibSp | Parch | Ticket | Fare | Cabin | Embarked | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 60 | 61 | 0 | 3 | Sirayanian, Mr. Orsen | male | 22.0 | 0 | 0 | 2669 | 7.2292 | NaN | C |
| 61 | 62 | 1 | 1 | Icard, Miss. Amelie | female | 38.0 | 0 | 0 | 113572 | 80.0000 | B28 | NaN |
| 62 | 63 | 0 | 1 | Harris, Mr. Henry Birkhardt | male | 45.0 | 1 | 0 | 36973 | 83.4750 | C83 | S |
| 63 | 64 | 0 | 3 | Skoog, Master. Harald | male | 4.0 | 3 | 2 | 347088 | 27.9000 | NaN | S |
| 64 | 65 | 0 | 1 | Stewart, Mr. Albert A | male | NaN | 0 | 0 | PC 17605 | 27.7208 | NaN | C |
def weighted_hamming(data):
""" Compute weighted hamming distance on categorical variables. For one variable, it is equal to 1 if
the values between point A and point B are different, else it is equal the relative frequency of the
distribution of the value across the variable. For multiple variables, the harmonic mean is computed
up to a constant factor.
@params:
- data = a pandas data frame of categorical variables
@returns:
- distance_matrix = a distance matrix with pairwise distance for all attributes
"""
categories_dist = []
for category in data:
X = pd.get_dummies(data[category])
X_mean = X * X.mean()
X_dot = X_mean.dot(X.transpose())
X_np = np.asarray(X_dot.replace(0,1,inplace=False))
categories_dist.append(X_np)
categories_dist = np.array(categories_dist)
distances = hmean(categories_dist, axis=0)
return distances
Weighted Hammintun 거리
data_categorical = ['Embarked']
train[data_categorical].shape
(891, 1)
X = pd.get_dummies(train['Embarked'])
X[60:65] ## 61 은 그냥, NAN 이란 그룹으로 남았다. 현재 따로 함수로 사용하기 때문에 이렇게 된 것이지. 실제로는 다른 값으로 missing value가 채워진다.
| C | Q | S | |
|---|---|---|---|
| 60 | 1 | 0 | 0 |
| 61 | 0 | 0 | 0 |
| 62 | 0 | 0 | 1 |
| 63 | 0 | 0 | 1 |
| 64 | 1 | 0 | 0 |
X = pd.get_dummies(train['Embarked'])
print(X.shape,'\n',X.iloc[60:65]) ## 카테고리밸류값이 3개이니, 3개의 컬럼을 가지게 된다.
X_mean = X * X.mean() ## dataframe*series 로, series의 인덱스, C,Q,R 의 평균값을 각각의 열별에 맞추어서 구하게 된다.
X_dot = X_mean.dot(X.transpose()) ## 자기의 Transformation 을 dot 하게 되면, n by n 정방행렬이 된다.
print(X_dot.iloc[60:65,0:10])
(891, 3)
C Q S
60 1 0 0
61 0 0 0
62 0 0 1
63 0 0 1
64 1 0 0
0 1 2 3 4 5 6 7 \
60 0.000000 0.188552 0.000000 0.000000 0.000000 0.0 0.000000 0.000000
61 0.000000 0.000000 0.000000 0.000000 0.000000 0.0 0.000000 0.000000
62 0.722783 0.000000 0.722783 0.722783 0.722783 0.0 0.722783 0.722783
63 0.722783 0.000000 0.722783 0.722783 0.722783 0.0 0.722783 0.722783
64 0.000000 0.188552 0.000000 0.000000 0.000000 0.0 0.000000 0.000000
8 9
60 0.000000 0.188552
61 0.000000 0.000000
62 0.722783 0.000000
63 0.722783 0.000000
64 0.000000 0.188552
X_np = np.asarray(X_dot.replace(0,1,inplace=False)) ## 0 인 값들을 1로 변환해준다.
print(X_np[60:65,0:10])
[[1. 0.18855219 1. 1. 1. 1.
1. 1. 1. 0.18855219]
[1. 1. 1. 1. 1. 1.
1. 1. 1. 1. ]
[0.72278339 1. 0.72278339 0.72278339 0.72278339 1.
0.72278339 0.72278339 0.72278339 1. ]
[0.72278339 1. 0.72278339 0.72278339 0.72278339 1.
0.72278339 0.72278339 0.72278339 1. ]
[1. 0.18855219 1. 1. 1. 1.
1. 1. 1. 0.18855219]]
categories_dist = []
categories_dist.append(X_np)
categories_dist = np.array(categories_dist)
distances = hmean(categories_dist, axis=0) ## Calculates the harmonic mean along the specified axis.
해석하면, 60번째 행과 거리가 가장 가까운 곳은 0.18855~ 값을 가지는 1번째,9번재 값을 보자.
값을 보게 되면, 아래와 같이 Embarked 값이 C 로 같은 것을 알 수 있다. 당연하게도, Embarked 값이 같기 때문이며, 0.18855~ 의 distance 값을 가지는 것은 모두 C 임을 알수있다.
train.iloc[[1,9,60],:]
| PassengerId | Survived | Pclass | Name | Sex | Age | SibSp | Parch | Ticket | Fare | Cabin | Embarked | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 1 | 1 | Cumings, Mrs. John Bradley (Florence Briggs Th... | female | 38.0 | 1 | 0 | PC 17599 | 71.2833 | C85 | C |
| 9 | 10 | 1 | 2 | Nasser, Mrs. Nicholas (Adele Achem) | female | 14.0 | 1 | 0 | 237736 | 30.0708 | NaN | C |
| 60 | 61 | 0 | 3 | Sirayanian, Mr. Orsen | male | 22.0 | 0 | 0 | 2669 | 7.2292 | NaN | C |
value_counts() 결과를 보면, S의 비율이 가장높고, 그 다음이 C , Q 임을 알 수 있다. 이런 빈도수 역시, 영향을 미치게 되는데, 상대적으로 많은 수를 가지는
X.mean()
C 0.188552
Q 0.086420
S 0.722783
dtype: float64
train.Embarked.value_counts() ##
S 644
C 168
Q 77
Name: Embarked, dtype: int64
하기 distance_matrix 함수가 사실 distance를 구하는 메인함수인데, weighted_hamming 용으로 상기 함수를 따로 빼둔것이다.
distance_matrix함수 역시, distance matrix 를 return한다.
def distance_matrix(data, numeric_distance = "euclidean", categorical_distance = "jaccard"): ## jaccard 개념은 따로 밑에서 부연한다.
""" Compute the pairwise distance attribute by attribute in order to account for different variables type:
- Continuous
- Categorical
For ordinal values, provide a numerical representation taking the order into account.
Categorical variables are transformed into a set of binary ones.
If both continuous and categorical distance are provided, a Gower-like distance is computed and the numeric
variables are all normalized in the process.
If there are missing values, the mean is computed for numerical attributes and the mode for categorical ones.
Note: If weighted-hamming distance is chosen, the computation time increases a lot since it is not coded in C
like other distance metrics provided by scipy.
@params:
- data = pandas dataframe to compute distances on.
- numeric_distances = the metric to apply to continuous attributes.
"euclidean" and "cityblock" available.
Default = "euclidean"
- categorical_distances = the metric to apply to binary attributes.
"jaccard", "hamming", "weighted-hamming" and "euclidean"
available. Default = "jaccard"
@returns:
- the distance matrix
"""
possible_continuous_distances = ["euclidean", "cityblock"]
possible_binary_distances = ["euclidean", "jaccard", "hamming", "weighted-hamming"]
number_of_variables = data.shape[1]
number_of_observations = data.shape[0]
# Get the type of each attribute (Numeric or categorical)
is_numeric = [all(isinstance(n, numbers.Number) for n in data.iloc[:, i]) for i, x in enumerate(data)]
is_all_numeric = sum(is_numeric) == len(is_numeric)
is_all_categorical = sum(is_numeric) == 0
is_mixed_type = not is_all_categorical and not is_all_numeric
# Check the content of the distances parameter
if numeric_distance not in possible_continuous_distances:
print("The continuous distance " + numeric_distance + " is not supported.")
return None
elif categorical_distance not in possible_binary_distances:
print("The binary distance " + categorical_distance + " is not supported.")
return None
# Separate the data frame into categorical and numeric attributes and normalize numeric data
if is_mixed_type:
number_of_numeric_var = sum(is_numeric)
number_of_categorical_var = number_of_variables - number_of_numeric_var
data_numeric = data.iloc[:, is_numeric]
data_numeric = (data_numeric - data_numeric.mean()) / (data_numeric.max() - data_numeric.min())
data_categorical = data.iloc[:, [not x for x in is_numeric]]
# Replace missing values with column mean for numeric values and mode for categorical ones. With the mode, it
# triggers a warning: "SettingWithCopyWarning: A value is trying to be set on a copy of a slice from a DataFrame"
# but the value are properly replaced
## 카테고릴 데이터의 missing value 는 일단, 맨 첫번째 mode()값으로 대채한다.
if is_mixed_type:
data_numeric.fillna(data_numeric.mean(), inplace=True) ## numeric 변수는 일단 mean() missing value를 대체한다.
for x in data_categorical:
data_categorical[x].fillna(data_categorical[x].mode()[0], inplace=True)
elif is_all_numeric:
data.fillna(data.mean(), inplace=True)
else:
for x in data:
data[x].fillna(data[x].mode()[0], inplace=True)
# "Dummifies" categorical variables in place
if not is_all_numeric and not (categorical_distance == 'hamming' or categorical_distance == 'weighted-hamming'):
if is_mixed_type:
data_categorical = pd.get_dummies(data_categorical)
else:
data = pd.get_dummies(data)
elif not is_all_numeric and categorical_distance == 'hamming':
if is_mixed_type:
data_categorical = pd.DataFrame([pd.factorize(data_categorical[x])[0] for x in data_categorical]).transpose()
else:
data = pd.DataFrame([pd.factorize(data[x])[0] for x in data]).transpose()
if is_all_numeric:
result_matrix = cdist(data, data, metric=numeric_distance)
elif is_all_categorical:
if categorical_distance == "weighted-hamming":
result_matrix = weighted_hamming(data)
else:
result_matrix = cdist(data, data, metric=categorical_distance)
else:
result_numeric = cdist(data_numeric, data_numeric, metric=numeric_distance)
if categorical_distance == "weighted-hamming":
result_categorical = weighted_hamming(data_categorical)
else:
result_categorical = cdist(data_categorical, data_categorical, metric=categorical_distance)
result_matrix = np.array([[1.0*(result_numeric[i, j] * number_of_numeric_var + result_categorical[i, j] *
number_of_categorical_var) / number_of_variables for j in range(number_of_observations)] for i in range(number_of_observations)])
# Fill the diagonal with NaN values
np.fill_diagonal(result_matrix, np.nan)
return pd.DataFrame(result_matrix)
print(train.shape)
train.iloc[60:65]
(891, 12)
| PassengerId | Survived | Pclass | Name | Sex | Age | SibSp | Parch | Ticket | Fare | Cabin | Embarked | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 60 | 61 | 0 | 3 | Sirayanian, Mr. Orsen | male | 22.0 | 0 | 0 | 2669 | 7.2292 | NaN | C |
| 61 | 62 | 1 | 1 | Icard, Miss. Amelie | female | 38.0 | 0 | 0 | 113572 | 80.0000 | B28 | NaN |
| 62 | 63 | 0 | 1 | Harris, Mr. Henry Birkhardt | male | 45.0 | 1 | 0 | 36973 | 83.4750 | C83 | S |
| 63 | 64 | 0 | 3 | Skoog, Master. Harald | male | 4.0 | 3 | 2 | 347088 | 27.9000 | NaN | S |
| 64 | 65 | 0 | 1 | Stewart, Mr. Albert A | male | NaN | 0 | 0 | PC 17605 | 27.7208 | NaN | C |
[‘Age’,’Cabin’,’Embarked’] 이 train 셋에서, null 값이 있다.
################sFunction distance_matrix 함수 탐구하기 Start################
tmp01 = [(i,x) for i, x in enumerate(train)] ## df 를 enumerate하면 컬럼을 순서데로 긁어온다.
print(tmp01)
[(0, 'PassengerId'), (1, 'Survived'), (2, 'Pclass'), (3, 'Name'), (4, 'Sex'), (5, 'Age'), (6, 'SibSp'), (7, 'Parch'), (8, 'Ticket'), (9, 'Fare'), (10, 'Cabin'), (11, 'Embarked')]
[all(isinstance(n, numbers.Number) for n in train.iloc[:, i]) for i, x in enumerate(train)]
[True, True, True, False, False, True, True, True, False, True, False, False]
train['Embarked'].mode()[0] ## model : 항상 Series를 return 하는 것. series에 [0]을 했으니, 첫번째 값이 나온다.
'S'
print(train['Embarked'].value_counts())
# print(train['Cabin'].value_counts())
print(len(pd.factorize(train['Embarked'])[0]))
pd.factorize(train['Embarked'])[0][0:10] ## R에서 factor 변수화 하는 것이랑 같다. label링으로 숫자형으로 바뀐다.
S 644
C 168
Q 77
Name: Embarked, dtype: int64
891
array([0, 1, 0, 0, 0, 2, 0, 0, 0, 1], dtype=int64)
data_categorical = ['Cabin','Embarked']
pd.DataFrame([pd.factorize(train[x])[0] for x in data_categorical])
| 0 | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | ... | 881 | 882 | 883 | 884 | 885 | 886 | 887 | 888 | 889 | 890 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | -1 | 0 | -1 | 1 | -1 | -1 | 2 | -1 | -1 | -1 | ... | -1 | -1 | -1 | -1 | -1 | -1 | 145 | -1 | 146 | -1 |
| 1 | 0 | 1 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 1 | ... | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 1 | 2 |
2 rows × 891 columns
a
| C | Q | S | |
|---|---|---|---|
| 0 | 0 | 0 | 1 |
| 1 | 1 | 0 | 0 |
| 2 | 0 | 0 | 1 |
| 3 | 0 | 0 | 1 |
| 4 | 0 | 0 | 1 |
a * a.mean()
| C | Q | S | |
|---|---|---|---|
| 0 | 0.0 | 0.0 | 0.8 |
| 1 | 0.2 | 0.0 | 0.0 |
| 2 | 0.0 | 0.0 | 0.8 |
| 3 | 0.0 | 0.0 | 0.8 |
| 4 | 0.0 | 0.0 | 0.8 |
categories_dist.shape
(1, 891, 891)
################sFunction distance_matrix 함수 탐구하기 End################
def knn_impute(target, attributes, k_neighbors, aggregation_method="mean", numeric_distance="euclidean",
categorical_distance="jaccard", missing_neighbors_threshold = 0.5):
""" Replace the missing values within the target variable based on its k nearest neighbors identified with the
attributes variables. If more than 50% of its neighbors are also missing values, the value is not modified and
remains missing. If there is a problem in the parameters provided, returns None.
If to many neighbors also have missing values, leave the missing value of interest unchanged.
@params:
- target = a vector of n values with missing values that you want to impute. The length has
to be at least n = 3.
- attributes = a data frame of attributes with n rows to match the target variable
- k_neighbors = the number of neighbors to look at to impute the missing values. It has to be a
value between 1 and n.
- aggregation_method = how to aggregate the values from the nearest neighbors (mean, median, mode)
Default = "mean"
- numeric_distances = the metric to apply to continuous attributes.
"euclidean" and "cityblock" available.
Default = "euclidean"
- categorical_distances = the metric to apply to binary attributes.
"jaccard", "hamming", "weighted-hamming" and "euclidean"
available. Default = "jaccard"
- missing_neighbors_threshold = minimum of neighbors among the k ones that are not also missing to infer
the correct value. Default = 0.5
@returns:
target_completed = the vector of target values with missing value replaced. If there is a problem
in the parameters, return None
"""
# Get useful variables
possible_aggregation_method = ["mean", "median", "mode"]
number_observations = len(target)
is_target_numeric = all(isinstance(n, numbers.Number) for n in target)
# Check for possible errors
if number_observations < 3:
print( "Not enough observations.")
return None
if attributes.shape[0] != number_observations:
print("The number of observations in the attributes variable is not matching the target variable length.")
return None
if k_neighbors > number_observations or k_neighbors < 1:
print("The range of the number of neighbors is incorrect.")
return None
if aggregation_method not in possible_aggregation_method:
print("The aggregation method is incorrect.")
return None
if not is_target_numeric and aggregation_method != "mode":
print("The only method allowed for categorical target variable is the mode.")
return None
# Make sure the data are in the right format
target = pd.DataFrame(target)
attributes = pd.DataFrame(attributes)
# Get the distance matrix and check whether no error was triggered when computing it
distances = distance_matrix(attributes, numeric_distance, categorical_distance) ## target 컬럼을 제외하고 distance를 구한다.
if distances is None:
return None
# Get the closest points and compute the correct aggregation method
for i, value in enumerate(target.iloc[:, 0]):
if pd.isnull(value):
order = distances.iloc[i,:].values.argsort()[:k_neighbors] ## argsort():오름차순으로의 원소들들의 index 값을 반환한다. 즉 거리가 가까운 index 들을 부른다.
closest_to_target = target.iloc[order, :]
# print("closest_to_target \n",closest_to_target)
# print(type(closest_to_target),closest_to_target.shape)
missing_neighbors = [x for x in closest_to_target.isnull().iloc[:, 0]]
# Compute the right aggregation method if at least more than 50% of the closest neighbors are not missing
if sum(missing_neighbors) >= missing_neighbors_threshold * k_neighbors:
continue
elif aggregation_method == "mean":
target.iloc[i] = np.ma.mean(np.ma.masked_array(closest_to_target,np.isnan(closest_to_target)))
elif aggregation_method == "median":
target.iloc[i] = np.ma.median(np.ma.masked_array(closest_to_target,np.isnan(closest_to_target)))
else: ## aggregation_method == "mode" 일때를 의미한다.
# target.iloc[i] = stats.mode(closest_to_target, nan_policy='omit')[0][0] ## stat 에 dataframe param을 받을수는 있으나, NaN 인식이 불량하다.
## closest_to_target 는 어짜피 dataFrame 이고, 이 구절은 na 을 없애는게 목적이니, 하기와 같이 대체한다.
closest_to_target.dropna(inplace=True)
target.iloc[i] = closest_to_target.iloc[0][0]
return target
knn_impute 실제 활용하기
Age null 값채우기 - Numeric 컬럼
print("before\n",train.loc[5:10,'Age'])
before
5 NaN
6 54.0
7 2.0
8 27.0
9 14.0
10 4.0
Name: Age, dtype: float64
new_train = knn_impute(target=train['Age'], attributes=train.drop(['Age', 'PassengerId'], 1),\
aggregation_method="median", k_neighbors=10, numeric_distance='euclidean',\
categorical_distance='hamming', missing_neighbors_threshold=0.8)
print(new_train.shape)
(891, 1)
print("after\n",new_train.loc[5:10,'Age'])
before
5 48.5
6 54.0
7 2.0
8 27.0
9 14.0
10 4.0
Name: Age, dtype: float64
Embarked null 값채우기 - Categorical 컬럼
print("before\n",train.loc[60:65,'Embarked'])
before
60 C
61 NaN
62 S
63 S
64 C
65 C
Name: Embarked, dtype: object
## data_categorical = ['Cabin','Embarked']
## ["euclidean", "jaccard", "hamming", "weighted-hamming"]
new_train_Embarked = knn_impute(target=train['Embarked'], attributes=train.drop(['Embarked', 'PassengerId'], 1),\
aggregation_method="mode", k_neighbors=10, numeric_distance='euclidean',\
categorical_distance='jaccard', missing_neighbors_threshold=0.8)
print("after\n",new_train_Embarked.loc[60:65,'Embarked'])
after
60 C
61 S
62 S
63 S
64 C
65 C
Name: Embarked, dtype: object
가장 가까운 이웃의 값으로 61번 행 값에, S가 들어왔음을 알 수 있다.

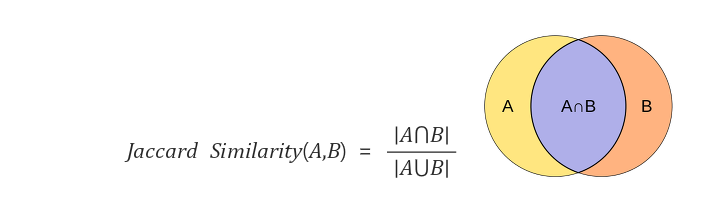

Comments