CatBoost Practice 01
Reference
- Medium Daniel Chepenko
- 상기 Blog 역자의 Collab 코드
Kaggle competition 에 있는 data를 활용한다.
보험회사에 공개한, 심각한 회사차원의 보험손실 여부를 추정한 regression 모델
import pandas as pd
df_train = pd.read_csv('../dataset/allstate-claims-severity/train.csv')
df_test = pd.read_csv('../dataset/allstate-claims-severity/test.csv')
import matplotlib.pyplot as plt
%matplotlib inline
import seaborn as sns
sns.set() # setting seaborn default for plots
print(df_train.shape) ## loss 값 있고,
print(df_test.shape) ## loss 값 없음.
(188318, 132)
(125546, 131)
df_train.head()
| id | cat1 | cat2 | cat3 | cat4 | cat5 | cat6 | cat7 | cat8 | cat9 | ... | cont6 | cont7 | cont8 | cont9 | cont10 | cont11 | cont12 | cont13 | cont14 | loss | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 1 | A | B | A | B | A | A | A | A | B | ... | 0.718367 | 0.335060 | 0.30260 | 0.67135 | 0.83510 | 0.569745 | 0.594646 | 0.822493 | 0.714843 | 2213.18 |
| 1 | 2 | A | B | A | A | A | A | A | A | B | ... | 0.438917 | 0.436585 | 0.60087 | 0.35127 | 0.43919 | 0.338312 | 0.366307 | 0.611431 | 0.304496 | 1283.60 |
| 2 | 5 | A | B | A | A | B | A | A | A | B | ... | 0.289648 | 0.315545 | 0.27320 | 0.26076 | 0.32446 | 0.381398 | 0.373424 | 0.195709 | 0.774425 | 3005.09 |
| 3 | 10 | B | B | A | B | A | A | A | A | B | ... | 0.440945 | 0.391128 | 0.31796 | 0.32128 | 0.44467 | 0.327915 | 0.321570 | 0.605077 | 0.602642 | 939.85 |
| 4 | 11 | A | B | A | B | A | A | A | A | B | ... | 0.178193 | 0.247408 | 0.24564 | 0.22089 | 0.21230 | 0.204687 | 0.202213 | 0.246011 | 0.432606 | 2763.85 |
5 rows × 132 columns
df_test.head(2)
| id | cat1 | cat2 | cat3 | cat4 | cat5 | cat6 | cat7 | cat8 | cat9 | ... | cont5 | cont6 | cont7 | cont8 | cont9 | cont10 | cont11 | cont12 | cont13 | cont14 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 4 | A | B | A | A | A | A | A | A | B | ... | 0.281143 | 0.466591 | 0.317681 | 0.61229 | 0.34365 | 0.38016 | 0.377724 | 0.369858 | 0.704052 | 0.392562 |
| 1 | 6 | A | B | A | B | A | A | A | A | B | ... | 0.836443 | 0.482425 | 0.443760 | 0.71330 | 0.51890 | 0.60401 | 0.689039 | 0.675759 | 0.453468 | 0.208045 |
2 rows × 131 columns
print("Number of missing values in train",df_train.isnull().sum().sum())
print("Number of missing values in test",df_test.isnull().sum().sum())
Number of missing values in train 0
Number of missing values in test 0
- 한꺼번에 전처리 하기 위해 묶은다음에 수행한다.
df_train_idx = df_train.index
df_test_idx = df_test.index
df_train['isTrain'] = True
df_test['isTrain'] = False
traintest = pd.concat([df_train, df_test], axis = 0)
set(df_train.columns)-set(df_test.columns)
{'loss'}
print(traintest.shape) ## 왜 133 이 되었지? -> isTrain 컬럼이 추가됨
(313864, 133)
- 컬럼이름 정리
import re
cat_pattern = re.compile("^cat([1-9]|[1-9][0-9]|[1-9][0-9][0-9])$")
cont_pattern = re.compile("^cont([1-9]|[1-9][0-9]|[1-9][0-9][0-9])$")
## cat이 들어간 컬럼. catogory 컬럼을 고르고, 뒤 3자리 숫자에 대해서, sorting
cat_col = sorted([cat for cat in traintest.columns if 'cat' in cat], key = lambda s: int(s[3:]))
## 정규식 패턴에 매칭되는 (위에서 구한 cat_col)들의 index 나열
cat_index = [i for i in range(0,len(traintest.columns)-1) if cat_pattern.match(traintest.columns[i])]
cont_col = sorted([cont for cont in traintest.columns if 'cont' in cont], key = lambda s: int(s[4:]))
cont_index = [i for i in range(0,len(traintest.columns)-1) if cont_pattern.match(traintest.columns[i])]
features = cat_col + cont_col
print(len(features)) ## 3개가 줄음 : isTrain,loss,id 컬럼
feats_counts = traintest[cat_col].nunique(dropna = False)
130
1-1. EDA columns check
len(cat_col),len(cont_col)
(116, 14)
feats_counts.sort_values() ## 대략 categorical 컬럼의 cardinality 를 확인하기 위해 자주 사용한다.
cat1 2
cat52 2
cat51 2
cat50 2
cat49 2
...
cat112 51
cat113 63
cat109 85
cat110 134
cat116 349
Length: 116, dtype: int64
## .nunique : return pandas.core.series.Series / 각컬럼값들의 unique한 갯수를 확인할 수 있음.
nunique = df_train[cat_col].nunique(dropna = False)
print(len(nunique))
nunique[100:110]
116
cat101 19
cat102 9
cat103 13
cat104 17
cat105 20
cat106 17
cat107 20
cat108 11
cat109 84
cat110 131
dtype: int64
plt.figure(figsize=(14,6))
_ = plt.hist(nunique, bins=100)
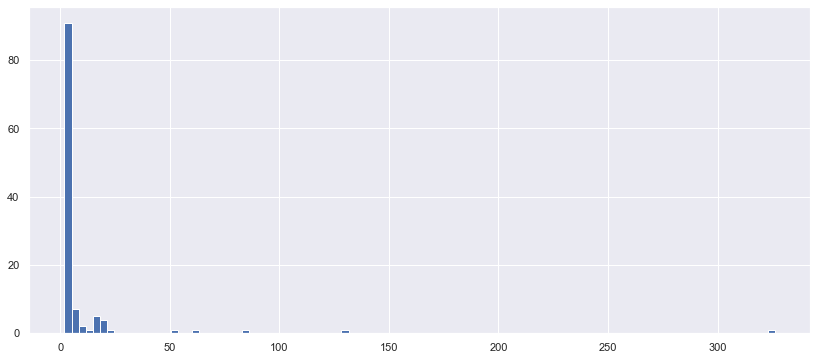
대부분의 cardinality가 10 개 이하의, 2개 짜리의 컬럼이 매우 많고. cardinality 300 이 넘는 것도 있다.
mask = (nunique > 100) ## cardinality 100 개 넘는 column 들만, 따로 dataframe 으로 빼서 확인하기
df_train[cat_col].loc[:, mask]
## cat110,cat116 컬럼의 cardinality가 100 이 넘음
| cat110 | cat116 | |
|---|---|---|
| 0 | BC | LB |
| 1 | CQ | DP |
| 2 | DK | GK |
| 3 | CS | DJ |
| 4 | C | CK |
| ... | ... | ... |
| 188313 | EG | CK |
| 188314 | BT | DF |
| 188315 | DM | DJ |
| 188316 | AI | MD |
| 188317 | EG | MJ |
188318 rows × 2 columns
df_train.cat110.value_counts()
CL 25305
EG 24654
CS 24592
EB 21396
CO 17495
...
BI 1
BM 1
S 1
DV 1
CB 1
Name: cat110, Length: 131, dtype: int64
상기 같이 cardinality 높은 컬럼들은 뉴메릭으로 변환하거나, zero variance groups 으로 이니 제거하는게 나을 수 있다.
df_train.id.is_unique
True
## cat110 컬럼에 대해 각 분류값별로, 얼마나 row 가 있는지 확인함 = df_train.cat110.value_counts()
## df_train.groupby('cat110')['id'].size().sort_values() 과 동일
cat110_nunique = df_train.groupby('cat110')['id'].nunique().sort_values(ascending=False)
cat110_nunique
cat110
CL 25305
EG 24654
CS 24592
EB 21396
CO 17495
...
DV 1
BD 1
BI 1
EH 1
BK 1
Name: id, Length: 131, dtype: int64
plt.figure(figsize=(14,6))
_ = plt.hist(cat110_nunique, bins=50)
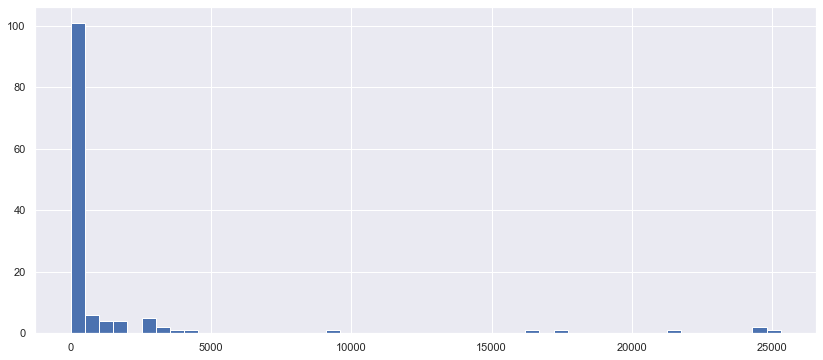
대부분 1~2개씩 있는 것으로 보인다.
cat116_nunique = df_train.groupby('cat116')['id'].nunique().sort_values()
plt.figure(figsize=(14,6))
_ = plt.hist(cat116_nunique, bins=50)
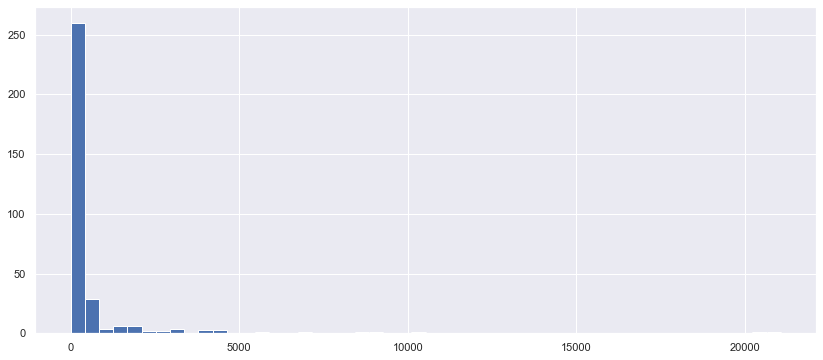
The values are not float, they are integer, so these features are likely to be even counts. Let’s look at another pack of features.
값은 부동하지 않고 정수이므로 이러한 특성은 짝수 카운트일 가능성이 높다. 다른 특징들을 살펴봅시다.
from tqdm.notebook import tqdm
df_train[cat_col].head()
| cat1 | cat2 | cat3 | cat4 | cat5 | cat6 | cat7 | cat8 | cat9 | cat10 | ... | cat107 | cat108 | cat109 | cat110 | cat111 | cat112 | cat113 | cat114 | cat115 | cat116 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | A | B | A | B | A | A | A | A | B | A | ... | J | G | BU | BC | C | AS | S | A | O | LB |
| 1 | A | B | A | A | A | A | A | A | B | B | ... | K | K | BI | CQ | A | AV | BM | A | O | DP |
| 2 | A | B | A | A | B | A | A | A | B | B | ... | F | A | AB | DK | A | C | AF | A | I | GK |
| 3 | B | B | A | B | A | A | A | A | B | A | ... | K | K | BI | CS | C | N | AE | A | O | DJ |
| 4 | A | B | A | B | A | A | A | A | B | B | ... | G | B | H | C | C | Y | BM | A | K | CK |
5 rows × 116 columns
df_train.cat1.unique().tolist()
['A', 'B']
df_train['cat112'].loc[df_train['cat112'].isin(df_train.cat1.unique().tolist())]
17 A
34 A
77 A
107 A
292 B
..
187955 A
188037 A
188266 A
188281 A
188303 B
Name: cat112, Length: 2834, dtype: object
## 예시
print(len(set(traintest['cat116'].factorize()[0])))
349
#Encoding categorical features to find duplicates
train_enc = pd.DataFrame(index = traintest.index)
## categorical = cat_col 에 대해서, 모두 숫자화로 변형시킨다. traintest = train+test concat
for col in tqdm(traintest[cat_col].columns):
train_enc[col] = traintest[col].factorize()[0]
HBox(children=(FloatProgress(value=0.0, max=116.0), HTML(value='')))
train_enc.head(3)
| cat1 | cat2 | cat3 | cat4 | cat5 | cat6 | cat7 | cat8 | cat9 | cat10 | ... | cat107 | cat108 | cat109 | cat110 | cat111 | cat112 | cat113 | cat114 | cat115 | cat116 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ... | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | ... | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 1 |
| 2 | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | ... | 2 | 2 | 2 | 2 | 1 | 2 | 2 | 0 | 1 | 2 |
3 rows × 116 columns
dup_cols = {} ## 중복되는 컬럼을 제거하는 과정
import numpy as np
for i, c1 in enumerate(tqdm(train_enc.columns)):
for c2 in train_enc.columns[i + 1:]: ## i번째 컬럼이후 다른 모든 컬럼
if c2 not in dup_cols and np.all(train_enc[c1] == train_enc[c2]): ## 바로 next 컬럼이, 지금 컬럼과 통으로 일치하면~
dup_cols[c2] = c1
HBox(children=(FloatProgress(value=0.0, max=116.0), HTML(value='')))
## 다행히 중복되는 컬럼은 없다.
dup_cols
{}
여기서, cat_col 에 대해 보면,, 모든 값이 알파벳의 조합으로만 되어있다. 따라서,
Factorize 하면, 결국 같은 value 들은 같은 숫자를 가지게 된다.
원작자는 이를 통해, 각 cat_columns 값이 같은면 1, 아니면 0으로 하여 평균값을 구하였고
이를 통해 컬럼들의 유사성을 파악했다.
test_list = ['a','b','c','d','e']
alst = []
for i,c1 in enumerate(test_list):
blst = []
for j,c2 in enumerate(test_list):
if i>=j: ##
blst.append(c1+c2)
# blst.append((train.loc[mask,c1].values>=train.loc[mask,c2].values).mean())
else: ## i < j 인 경우
blst.append(c2+c1)
# blst.append((train.loc[mask,c1].values>train.loc[mask,c2].values).mean())
alst.append(blst)
alst
[['aa', 'ba', 'ca', 'da', 'ea'],
['ba', 'bb', 'cb', 'db', 'eb'],
['ca', 'cb', 'cc', 'dc', 'ec'],
['da', 'db', 'dc', 'dd', 'ed'],
['ea', 'eb', 'ec', 'ed', 'ee']]
1-2. EDA 컬럼별 연관성 분석
cat_col들의 연관성 보는 함수 (위의 설명 참조)
def autolabel(arrayA):
''' label each colored square with the corresponding data value.
If value > 20, the text is in black, else in white.
'''
arrayA = np.array(arrayA)
for i in range(arrayA.shape[0]):
for j in range(arrayA.shape[1]):
plt.text(j,i, "%.2f"%arrayA[i,j], ha='center', va='bottom',color='w')
def gt_matrix(train, feats,sz=18): ## feats 컬럼명
a = []
for i,c1 in enumerate(feats):
b = []
for j,c2 in enumerate(feats):
mask = (~train[c1].isnull()) & (~train[c2].isnull()) ## c1,c2 컬럼 둘다 null 아닌 row 만 표시
if i>=j: ##
b.append((train.loc[mask,c1].values>=train.loc[mask,c2].values).mean())
else: ## i < j 인 경우
b.append((train.loc[mask,c1].values>train.loc[mask,c2].values).mean())
a.append(b)
plt.figure(figsize = (sz,sz))
plt.imshow(a, interpolation = 'None')
_ = plt.xticks(range(len(feats)),feats,rotation = 90)
_ = plt.yticks(range(len(feats)),feats,rotation = 0)
autolabel(a)
mask = (~train_enc['cat3'].isnull()) & (~train_enc['cat1'].isnull())
train_enc.loc[mask,'cat3'].values
array([0, 0, 0, ..., 0, 0, 0], dtype=int64)
## 대각 아래 영역 : 같거나, 큰거...
print(train_enc.loc[mask,'cat2'].values >= train_enc.loc[mask,'cat1'].values)
print((train_enc.loc[mask,'cat2'].values >= train_enc.loc[mask,'cat1'].values).mean())
[ True True True ... False True True]
0.9280388958274922
## 대각 상단 영역
print(train_enc.loc[mask,'cat1'].values > train_enc.loc[mask,'cat3'].values)
print((train_enc.loc[mask,'cat1'].values > train_enc.loc[mask,'cat3'].values).mean())
[False False False ... True False False]
0.24539609512400276
#from cat13 to cat30
gt_matrix(train_enc, cat_col[:30])
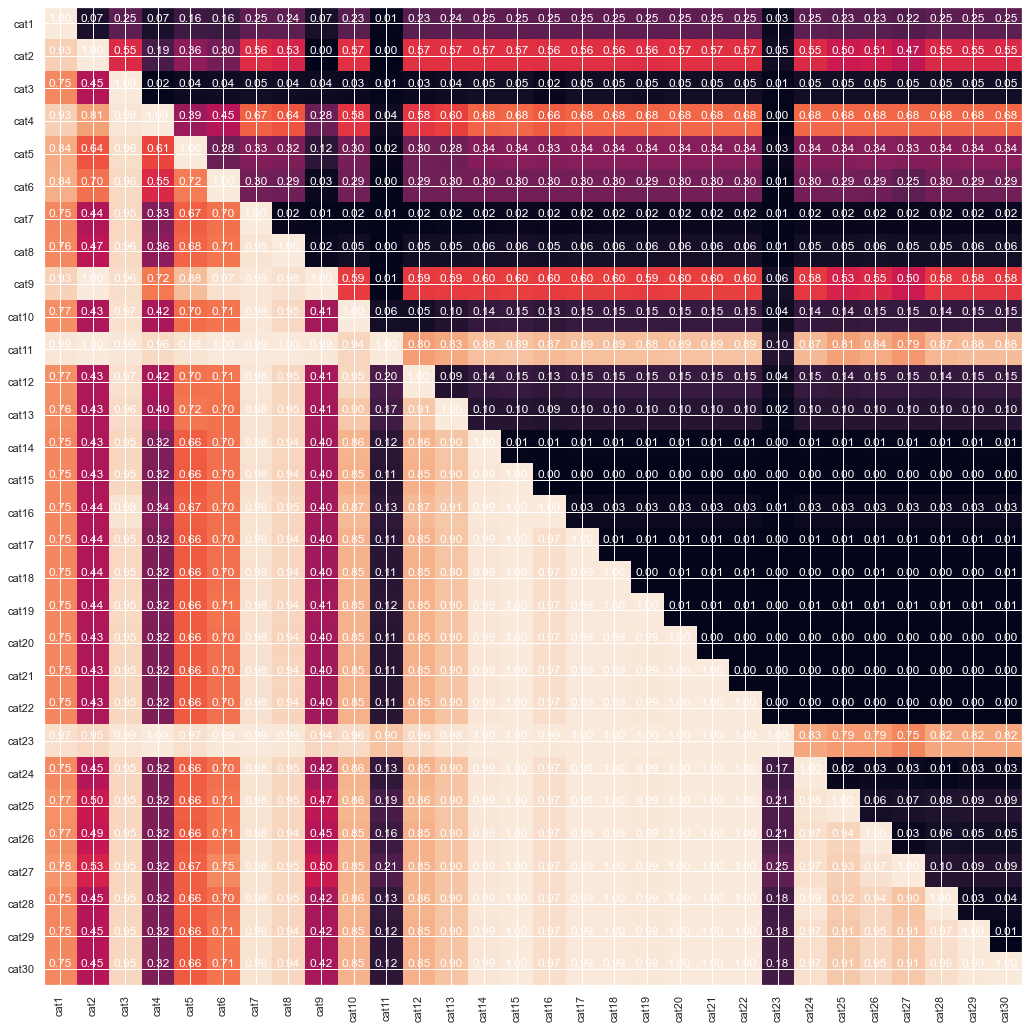
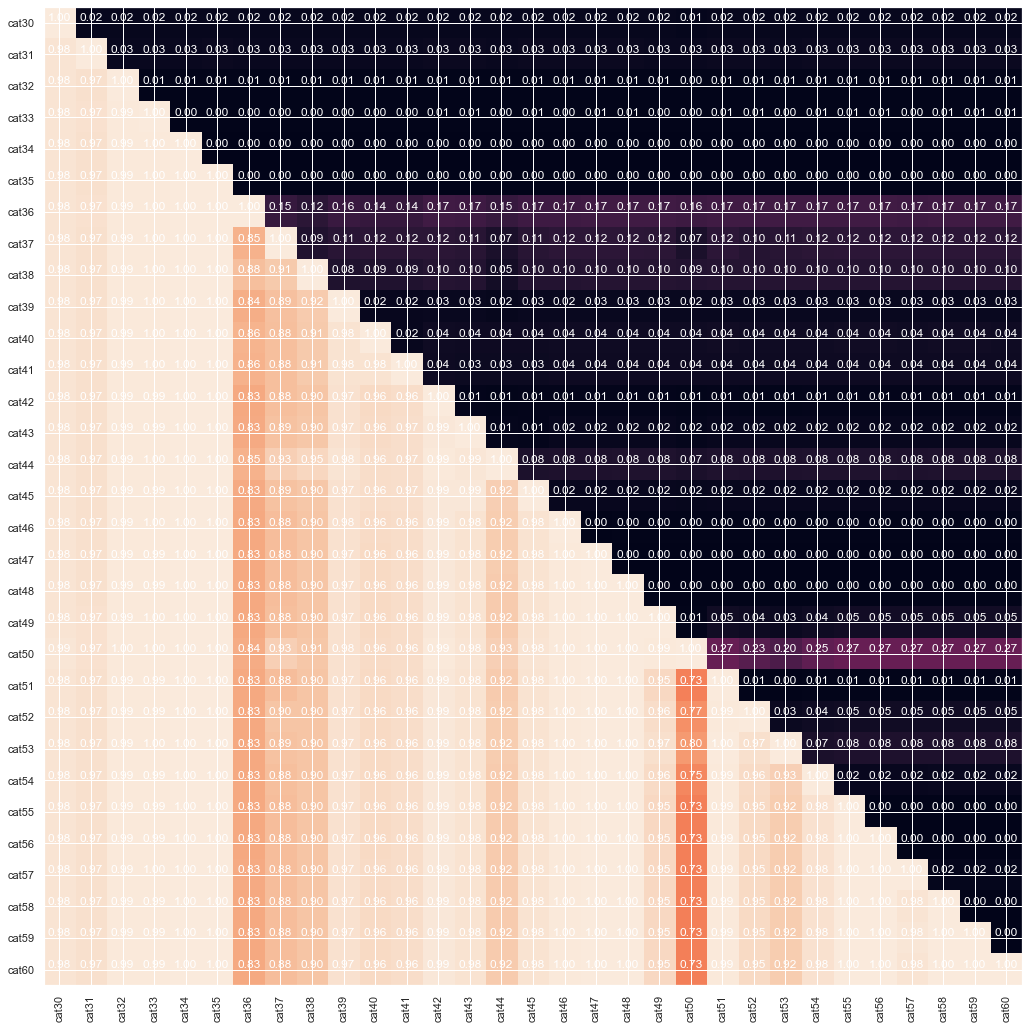
#everything except cat36, cat 37, cat50
gt_matrix(train_enc, cat_col[29:60])
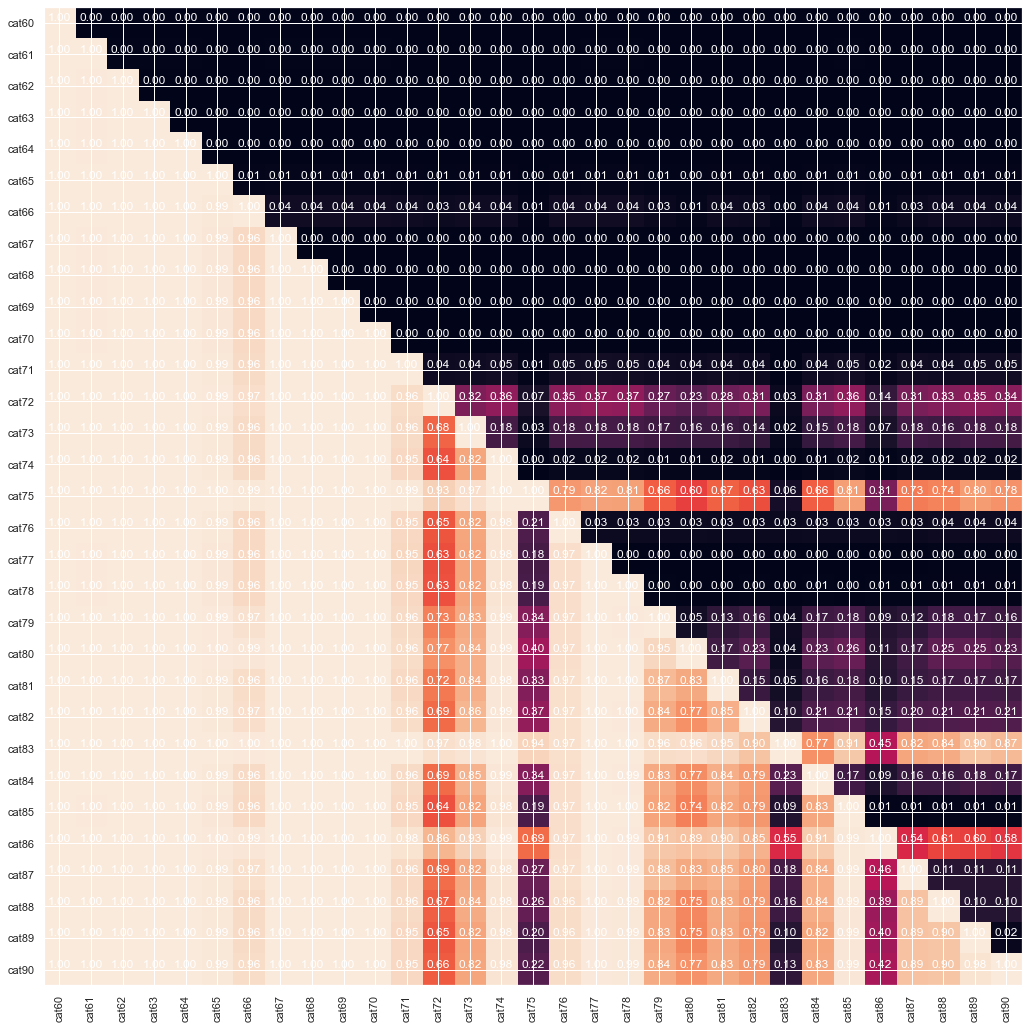
#from cat60 to cat71, cat74, from cat76 to cat78
gt_matrix(train_enc, cat_col[59:90])
cum_cat_1 = []
for i in range(12,23):
cum_cat_1.append('cat'+str(i))
cum_cat_2 = []
for i in range(24,50):
cum_cat_2.append('cat'+str(i))
cum_cat_3 = []
for i in range(51,72):
cum_cat_3.append('cat'+str(i))
From the graph above we can see that some of the features are cummulative, as in some cases the one is strictly greater than the other. Finally lets look at correlation between continious features. Probably dimension reduction techniques can help us
1-3. EDA -Numeric 변수들 상관계수 확인하기
threshold = 0.5
data_corr = traintest[cont_col].corr()
# List of pairs along with correlation above threshold
corr_list = []
#Search for the highly correlated pairs
for i in range(0,len(cont_col)): #for 'size' features
for j in range(i+1,len(cont_col)): #avoid repetition
if (data_corr.iloc[i,j] >= threshold and data_corr.iloc[i,j] < 1) or (data_corr.iloc[i,j] < 0 and data_corr.iloc[i,j] <= -threshold):
corr_list.append([data_corr.iloc[i,j],i,j]) #store correlation and columns index
#Sort to show higher ones first
s_corr_list = sorted(corr_list,key=lambda x: abs(x[0]),reverse=True)
#Print correlations and column names
for v,i,j in s_corr_list:
print ("%s and %s = %.2f" % (cont_col[i],cont_col[j],v))
cont11 and cont12 = 0.99
cont1 and cont9 = 0.93
cont6 and cont10 = 0.88
cont6 and cont13 = 0.81
cont1 and cont10 = 0.81
cont6 and cont9 = 0.80
cont6 and cont12 = 0.79
cont9 and cont10 = 0.79
cont6 and cont11 = 0.77
cont1 and cont6 = 0.76
cont7 and cont11 = 0.75
cont7 and cont12 = 0.74
cont10 and cont12 = 0.72
cont10 and cont13 = 0.71
cont10 and cont11 = 0.70
cont6 and cont7 = 0.66
cont9 and cont13 = 0.64
cont9 and cont12 = 0.63
cont1 and cont12 = 0.61
cont9 and cont11 = 0.61
cont1 and cont11 = 0.60
cont1 and cont13 = 0.53
cont4 and cont8 = 0.53
1-4. EDA - PCA dimension reduction
from sklearn.preprocessing import StandardScaler
from sklearn.decomposition import PCA
traintest.shape[0] == len(df_train_idx.to_list())+len(df_test_idx.to_list())
True
If 0 < n_components < 1 and svd_solver == ‘full’
select the number of components such that the amount of variance that needs to be explained is greater than the percentage specified by n_components.
diag 고유값행렬 (eigen value (lambda)) 의 0.95(95%)에 해당하는 변수들을 모두 본다는 의미임
만약 0.95 -> 0.7 -> 0.5 로 될 수록, 백분위수 컨셉으로 총량의 variance 가 작아지니, len(pca.explained_variance_) 이 작아진다.
## numeric 변수들로만 시행
scaler = StandardScaler()
traintest_scaled = scaler.fit_transform(traintest[cont_col]) ## modify scale in numeric columns
print(traintest_scaled.shape)
(313864, 14)
pca = PCA(n_components=0.95, svd_solver='full').fit(traintest_scaled)
traintest_pca = pca.transform(traintest_scaled)
print(len(pca.explained_variance_)),print(pca.explained_variance_)
9
[6.15481809 2.00696816 1.49785047 0.99611156 0.88174836 0.75109768
0.56965726 0.39493657 0.30094088]
(None, None)
traintest_pca_df = pd.DataFrame(data = traintest_pca)
traintest_pca_df['id'] = traintest['id'].values
print(traintest[cont_col].shape)
(313864, 14)
np.dot(traintest_scaled,pca.components_.T)[0]
array([ 2.70453183, -2.10166972, -1.51128802, 0.61746921, -0.54985965,
-0.21919071, 0.94667346, 1.10674658, -1.36947719])
print(traintest_pca_df.shape)
traintest_pca_df.head()
(313864, 10)
| 0 | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | id | |
|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 2.704532 | -2.101670 | -1.511288 | 0.617469 | -0.549860 | -0.219191 | 0.946673 | 1.106747 | -1.369477 | 1 |
| 1 | -1.132381 | -0.395118 | 2.283496 | -0.762693 | -0.352918 | -0.593252 | -1.064844 | 0.443155 | 0.220732 | 2 |
| 2 | -2.948887 | 0.339230 | -1.375522 | 1.256290 | -0.212120 | 0.107400 | 0.040797 | 0.128152 | -0.309722 | 5 |
| 3 | -1.597690 | 0.072182 | -0.424028 | 0.392106 | -0.004357 | -1.252742 | -0.050669 | 0.638594 | -0.293227 | 10 |
| 4 | -4.068934 | -1.052458 | -0.660599 | -0.245261 | -0.841898 | 0.906329 | -0.748438 | 0.017867 | -0.517443 | 11 |
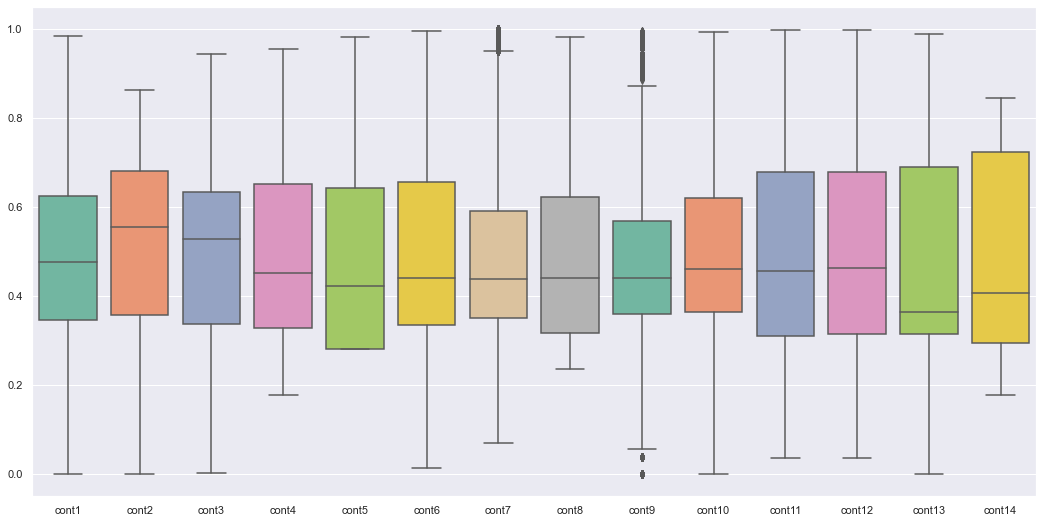
traintest[cont_col].describe()
| cont1 | cont2 | cont3 | cont4 | cont5 | cont6 | cont7 | cont8 | cont9 | cont10 | cont11 | cont12 | cont13 | cont14 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| count | 313864.000000 | 313864.000000 | 313864.000000 | 313864.000000 | 313864.000000 | 313864.000000 | 313864.000000 | 313864.000000 | 313864.00000 | 313864.000000 | 313864.000000 | 313864.000000 | 313864.000000 | 313864.000000 |
| mean | 0.494096 | 0.507089 | 0.498653 | 0.492021 | 0.487513 | 0.491442 | 0.485360 | 0.486823 | 0.48571 | 0.498403 | 0.493850 | 0.493503 | 0.493917 | 0.495665 |
| std | 0.187768 | 0.207056 | 0.201961 | 0.211101 | 0.209063 | 0.205394 | 0.178531 | 0.199442 | 0.18185 | 0.185906 | 0.210002 | 0.209716 | 0.212911 | 0.222537 |
| min | 0.000016 | 0.001149 | 0.002634 | 0.176921 | 0.281143 | 0.012683 | 0.069503 | 0.236880 | 0.00008 | 0.000000 | 0.035321 | 0.036232 | 0.000228 | 0.178568 |
| 25% | 0.347403 | 0.358319 | 0.336963 | 0.327354 | 0.281143 | 0.336105 | 0.351299 | 0.317960 | 0.35897 | 0.364580 | 0.310961 | 0.314945 | 0.315758 | 0.294657 |
| 50% | 0.475784 | 0.555782 | 0.527991 | 0.452887 | 0.422268 | 0.440945 | 0.438650 | 0.441060 | 0.44145 | 0.461190 | 0.457203 | 0.462286 | 0.363547 | 0.407020 |
| 75% | 0.625272 | 0.681761 | 0.634224 | 0.652072 | 0.643315 | 0.655818 | 0.591165 | 0.623580 | 0.56889 | 0.619840 | 0.678924 | 0.679096 | 0.689974 | 0.724707 |
| max | 0.984975 | 0.862654 | 0.944251 | 0.956046 | 0.983674 | 0.997162 | 1.000000 | 0.982800 | 0.99540 | 0.994980 | 0.998742 | 0.998484 | 0.988494 | 0.844848 |
plt.figure(figsize=(18,9))
ax = sns.boxplot(data=traintest[cont_col], orient="v", palette="Set2")
1-5. EDA - 데이터 왜도(skewness) 확인
For normally distributed data, the skewness should be about zero. For unimodal continuous distributions, a skewness value greater than zero means that there is more weight in the right tail of the distribution. The function skewtest can be used to determine if the skewness value is close enough to zero, statistically speaking.
from scipy.stats import skew ## Compute the sample skewness of a data set.
skewed_cols = []
for col in cont_col:
if skew(traintest[col]) > 0.75:
skewed_cols.append(col)
## 왜도가 0.75 넘는 컬럼 찾아내기
skewed_cols
['cont7', 'cont9']
for col in skewed_cols:
plt.figure()
plt.title(col)
plt.hist(traintest[col], bins = 50)
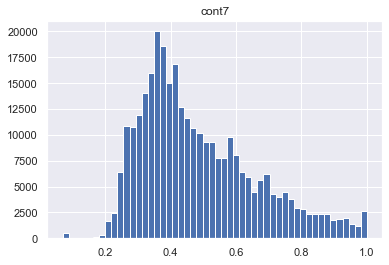
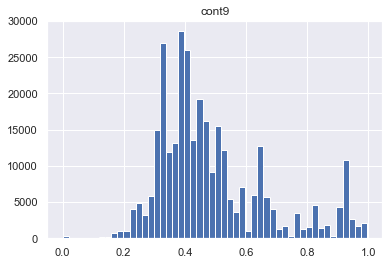
As we see the features cont7 and cont 9 are left skewed. So we need to apply transformation. BoxCox could be a good way to deal with this problem
from scipy import stats
stats.boxcox(traintest[col])[0] ## unnormal distribution data -> normal distribution data (일종의 Skill)
array([-0.37071516, -0.86886216, -1.06116485, ..., -0.05663025,
-0.92868009, -0.94375053])
for col in skewed_cols:
col_name = col+"_"
traintest[col_name] = stats.boxcox(traintest[col])[0]
plt.figure()
plt.title(col_name)
plt.hist(traintest[col_name], bins = 50)
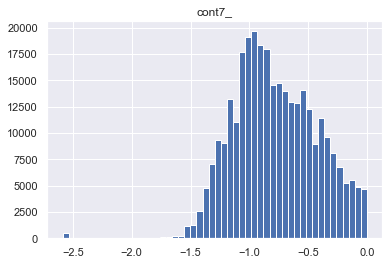
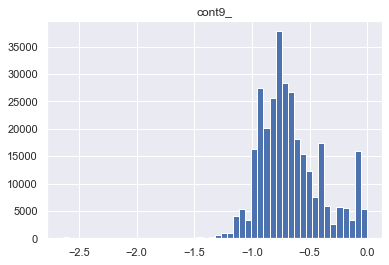
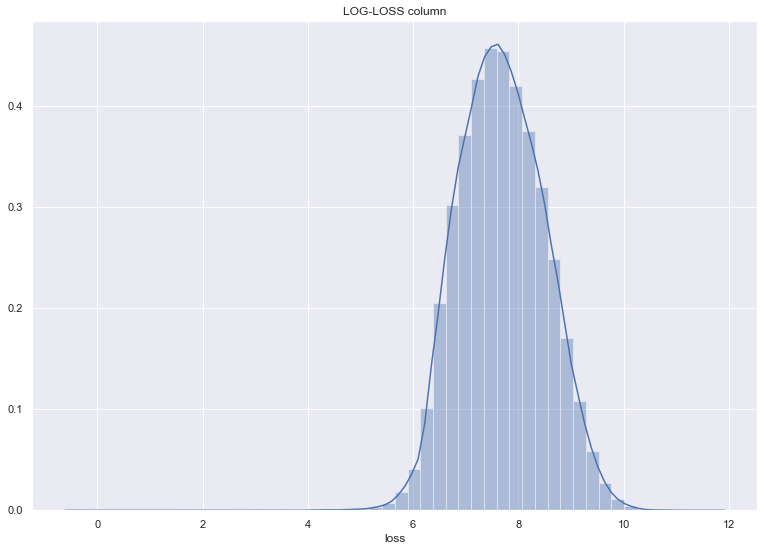
1-6. EDA - Y 컬럼(=loss) 컬럼의 밀도를 보고, left skewness 인것에 대해 log 변환한다.
## log 를 씌워서, loss 컬럼의 밀도 살펴보기
plt.figure(figsize=(13,9))
ax = sns.distplot(df_train["loss"])
ax.set_title('NO-LOG-LOSS column')
Text(0.5, 1.0, 'NO-LOG-LOSS column')
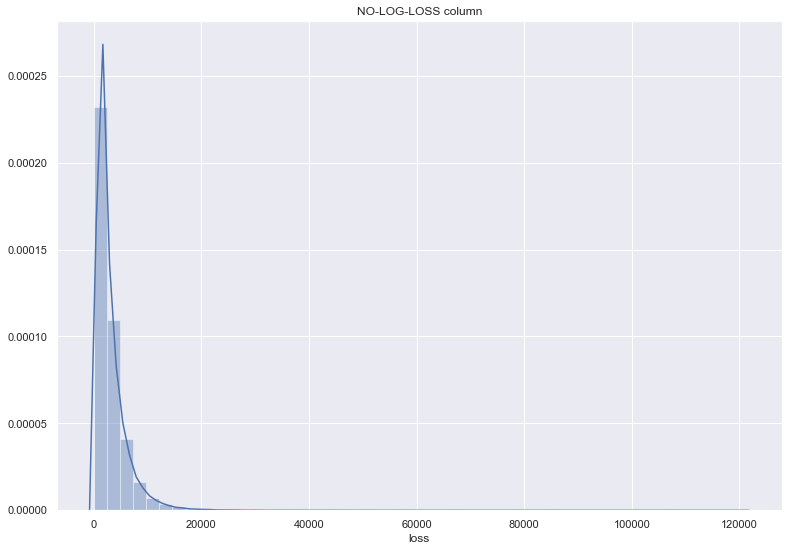
## log 를 씌워서, loss 컬럼의 밀도 살펴보기
plt.figure(figsize=(13,9))
sns.distplot(np.log(df_train["loss"])).set_title('LOG-LOSS column')
Text(0.5, 1.0, 'LOG-LOSS column')
2-1. Modeling Without tunning, apply Catboost - Base
- y 에 대해서는 np.log(y) 로 변형시켜서 사용했다.
- Firslty try to run model without any EDA transformation
from catboost import CatBoostRegressor
df_train.head(2)
| id | cat1 | cat2 | cat3 | cat4 | cat5 | cat6 | cat7 | cat8 | cat9 | ... | cont7 | cont8 | cont9 | cont10 | cont11 | cont12 | cont13 | cont14 | loss | isTrain | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 1 | A | B | A | B | A | A | A | A | B | ... | 0.335060 | 0.30260 | 0.67135 | 0.83510 | 0.569745 | 0.594646 | 0.822493 | 0.714843 | 2213.18 | True |
| 1 | 2 | A | B | A | A | A | A | A | A | B | ... | 0.436585 | 0.60087 | 0.35127 | 0.43919 | 0.338312 | 0.366307 | 0.611431 | 0.304496 | 1283.60 | True |
2 rows × 133 columns
df_train.isTrain.value_counts()
True 188318
Name: isTrain, dtype: int64
## features : loss,isTrain 컬럼을 제외한 순수 Feature 컬럼
X = df_train.drop('id', axis = 1)[features]
y = np.log(df_train['loss'])
from sklearn.model_selection import train_test_split
X_train, X_validation, y_train, y_validation = train_test_split(X, y, train_size=0.8)
## cat이 들어간 컬럼. catogory 컬럼을 고르고, 뒤 3자리 숫자에 대해서, sorting
cat_col = sorted([cat for cat in X_train.columns if 'cat' in cat], key = lambda s: int(s[3:]))
## 정규식 패턴에 매칭되는 (위에서 구한 cat_col)들의 index 나열
cat_index = [i for i in range(0,len(X_train.columns)-1) if cat_pattern.match(X_train.columns[i])]
cont_col = sorted([cont for cont in X_train.columns if 'cont' in cont], key = lambda s: int(s[4:]))
cont_index = [i for i in range(0,len(X_train.columns)-1) if cont_pattern.match(X_train.columns[i])]
features = cat_col + cont_col
params = {'iterations':100, 'learning_rate':0.1, 'eval_metric':"MAE"}
model_0 = CatBoostRegressor(**params)
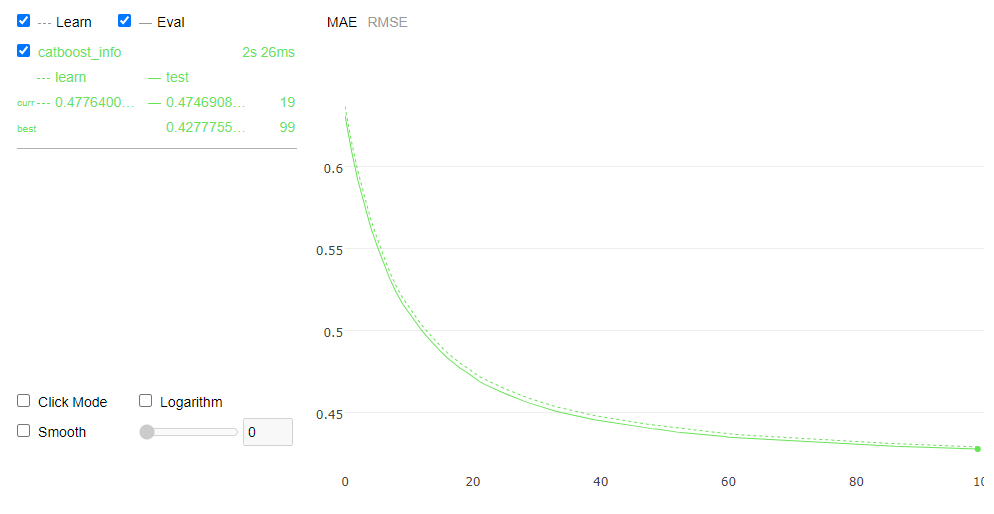
model_0.fit(X_train, y_train, cat_index, eval_set=(X_validation, y_validation) ,plot=True,verbose=False,use_best_model=True)
print(model_0.evals_result_.keys())
print(model_0.learning_rate_,model_0.tree_count_)
mae_min_idx = np.argmin(model_0.evals_result_['validation']['MAE'])
print("Minimize MAE value is {:.6f} in Validation Set".format(model_0.evals_result_['validation']['MAE'][mae_min_idx]))
dict_keys(['learn', 'validation'])
0.10000000149011612 100
Minimize MAE value is 0.427776 in Validation Set
## y 결과값에 log 변환한 값이 나올테니, 이를 다시 원본값으로 돌리기 위해 np.exp()를 씌웠다.
df_test['loss'] = np.exp(model_0.predict(df_test[features]))
df_test.head()
| id | cat1 | cat2 | cat3 | cat4 | cat5 | cat6 | cat7 | cat8 | cat9 | ... | cont7 | cont8 | cont9 | cont10 | cont11 | cont12 | cont13 | cont14 | isTrain | loss | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 4 | A | B | A | A | A | A | A | A | B | ... | 0.317681 | 0.61229 | 0.34365 | 0.38016 | 0.377724 | 0.369858 | 0.704052 | 0.392562 | False | 1597.451430 |
| 1 | 6 | A | B | A | B | A | A | A | A | B | ... | 0.443760 | 0.71330 | 0.51890 | 0.60401 | 0.689039 | 0.675759 | 0.453468 | 0.208045 | False | 1960.685945 |
| 2 | 9 | A | B | A | B | B | A | B | A | B | ... | 0.325779 | 0.29758 | 0.34365 | 0.30529 | 0.245410 | 0.241676 | 0.258586 | 0.297232 | False | 8500.643058 |
| 3 | 12 | A | A | A | A | B | A | A | A | A | ... | 0.342355 | 0.40028 | 0.33237 | 0.31480 | 0.348867 | 0.341872 | 0.592264 | 0.555955 | False | 5131.988609 |
| 4 | 15 | B | A | A | A | A | B | A | A | A | ... | 0.391833 | 0.23688 | 0.43731 | 0.50556 | 0.359572 | 0.352251 | 0.301535 | 0.825823 | False | 915.043904 |
5 rows × 133 columns
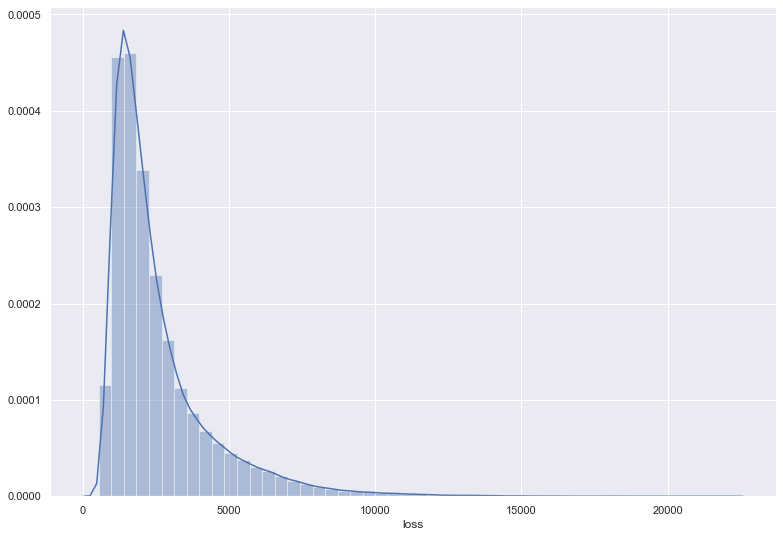
plt.figure(figsize=(13,9))
sns.distplot(df_test["loss"])
<matplotlib.axes._subplots.AxesSubplot at 0x151826c5448>
df_test.to_csv('../dataset/kaggle_submission/acs_submission_0.csv', sep = ',', columns = ['id', 'loss'], index=False)
2-2. Modiling With PCA, apply Catboost
- cont_col 만 PCA decomposition 시 사용했음. 주의할 것!
- Try to reduce dimenstions with PCA
traintest_pca_df.head()
| 0 | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | id | |
|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 2.704532 | -2.101670 | -1.511288 | 0.617469 | -0.549860 | -0.219191 | 0.946673 | 1.106747 | -1.369477 | 1 |
| 1 | -1.132381 | -0.395118 | 2.283496 | -0.762693 | -0.352918 | -0.593252 | -1.064844 | 0.443155 | 0.220732 | 2 |
| 2 | -2.948887 | 0.339230 | -1.375522 | 1.256290 | -0.212120 | 0.107400 | 0.040797 | 0.128152 | -0.309722 | 5 |
| 3 | -1.597690 | 0.072182 | -0.424028 | 0.392106 | -0.004357 | -1.252742 | -0.050669 | 0.638594 | -0.293227 | 10 |
| 4 | -4.068934 | -1.052458 | -0.660599 | -0.245261 | -0.841898 | 0.906329 | -0.748438 | 0.017867 | -0.517443 | 11 |
print("traintest_pca_df.shape :{}".format(traintest_pca_df.shape))
print("traintest.shape :{}".format(traintest.shape))
traintest_pca_df.shape :(313864, 10)
traintest.shape :(313864, 135)
df_train_idx,df_test_idx 각각에 해당하는 데이터를 골라냄 (index 를 이용해서)
traintest_pca_df 는 이미 PCA 를 이용해서,원래의 row,columns 등 shape가 축소된 형태임
type(df_train_idx)
pandas.core.indexes.range.RangeIndex
traintest_pca_df.loc[df_train_idx,:].shape
(188318, 10)
# df_train_pca = traintest_pca_df[traintest_pca_df['id'].isin(df_train_idx)]
# df_test_pca = traintest_pca_df[traintest_pca_df['id'].isin(df_test_idx)]
## 원본코드가 이상해서 바꿈
df_train_pca = traintest_pca_df.loc[df_train_idx,:]
df_test_pca = traintest_pca_df.loc[df_test_idx,:]
len(df_train_pca)
188318
print("df_train.shape :{}".format(df_train.shape))
print("df_test.shape :{}".format(df_test.shape))
print()
print("df_train_pca.shape :{}".format(df_train_pca.shape))
print("df_test_pca.shape :{}".format(df_test_pca.shape))
df_train.shape :(188318, 133)
df_test.shape :(125546, 133)
df_train_pca.shape :(188318, 10)
df_test_pca.shape :(125546, 10)
df_train_pca(pca 축소된 주요인 컬럼)을 id 기준으로 merge 함.</span>
이때, cont_col 컬럼들은 제외함(pca 가 cont_col 컬럼에 해당함으로)
정리하면, train_set 에서, cont 컬럼(뉴메릭컬럼) 은, id 값을 맞추어서, pca 로 추려진 요인들의 값으로 대체함
df_train_ = df_train.drop(cont_col, axis = 1).set_index('id').join(df_train_pca.set_index('id'))
df_test_ = df_test.drop(cont_col, axis = 1).set_index('id').join(df_test_pca.set_index('id'))
df_train_.head(2)
| cat1 | cat2 | cat3 | cat4 | cat5 | cat6 | cat7 | cat8 | cat9 | cat10 | ... | isTrain | 0 | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| id | |||||||||||||||||||||
| 1 | A | B | A | B | A | A | A | A | B | A | ... | True | 2.704532 | -2.101670 | -1.511288 | 0.617469 | -0.549860 | -0.219191 | 0.946673 | 1.106747 | -1.369477 |
| 2 | A | B | A | A | A | A | A | A | B | B | ... | True | -1.132381 | -0.395118 | 2.283496 | -0.762693 | -0.352918 | -0.593252 | -1.064844 | 0.443155 | 0.220732 |
2 rows × 127 columns
df_test_.head(2)
| cat1 | cat2 | cat3 | cat4 | cat5 | cat6 | cat7 | cat8 | cat9 | cat10 | ... | loss | 0 | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| id | |||||||||||||||||||||
| 4 | A | B | A | A | A | A | A | A | B | A | ... | 1597.451430 | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN |
| 6 | A | B | A | B | A | A | A | A | B | A | ... | 1960.685945 | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN |
2 rows × 127 columns
print("df_train_.shape :{}".format(df_train_.shape))
print("df_test_.shape :{}".format(df_test_.shape))
df_train_.shape :(188318, 127)
df_test_.shape :(125546, 127)
X = df_train_.drop('loss', axis = 1)
y = np.log(df_train_['loss'])
X_train, X_validation, y_train, y_validation = train_test_split(X, y, train_size=0.8)
model_1 = CatBoostRegressor(**params)
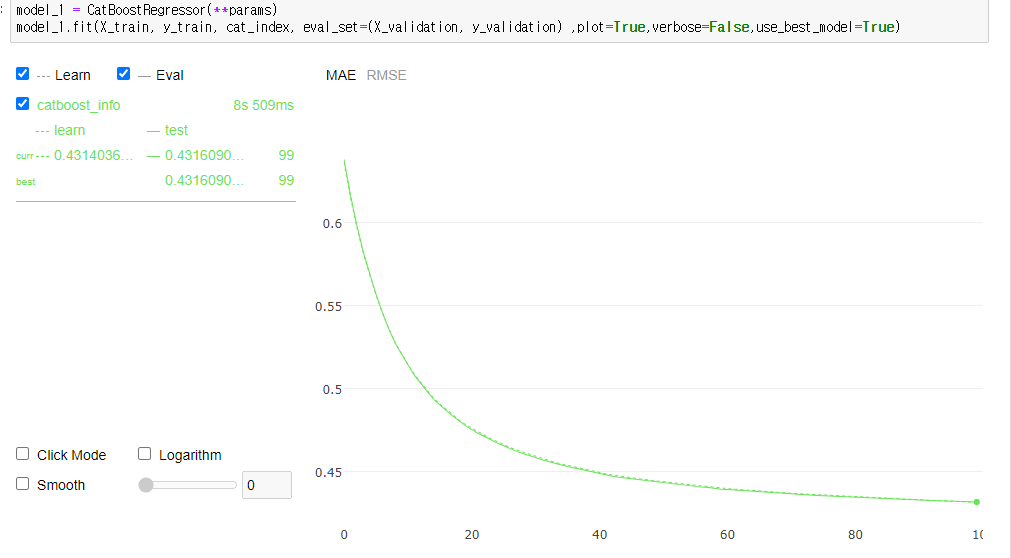
model_1.fit(X_train, y_train, cat_index, eval_set=(X_validation, y_validation) ,plot=True,verbose=False,use_best_model=True)
print(model_1.evals_result_.keys())
print(model_1.learning_rate_,model_1.tree_count_)
mae_min_idx = np.argmin(model_1.evals_result_['validation']['MAE'])
print("Minimize MAE value is {:.6f} in Validation Set".format(model_1.evals_result_['validation']['MAE'][mae_min_idx]))
dict_keys(['learn', 'validation'])
0.10000000149011612 100
Minimize MAE value is 0.431609 in Validation Set
## y 결과값에 log 변환한 값이 나올테니, 이를 다시 원본값으로 돌리기 위해 np.exp()를 씌웠다.
df_test_['loss'] = np.exp(model_1.predict(df_test_.drop('loss', axis=1)))
df_test_.to_csv('../dataset/kaggle_submission/acs_submission_1.csv', sep = ',', columns = ['loss'], index=False)
PCA를 활용한 model_1 을 tunning 하면,
# model_1.fit(X_train, y_train, cat_index, eval_set=(X_validation, y_validation) ,plot=True,verbose=False,use_best_model=True)
params
{'iterations': 100, 'learning_rate': 0.1, 'eval_metric': 'MAE'}
model_1_tuned = CatBoostRegressor(iterations=1500, learning_rate=0.05, depth =8, task_type = "GPU", eval_metric = "MAE", l2_leaf_reg=3, bagging_temperature=1,one_hot_max_size=0)
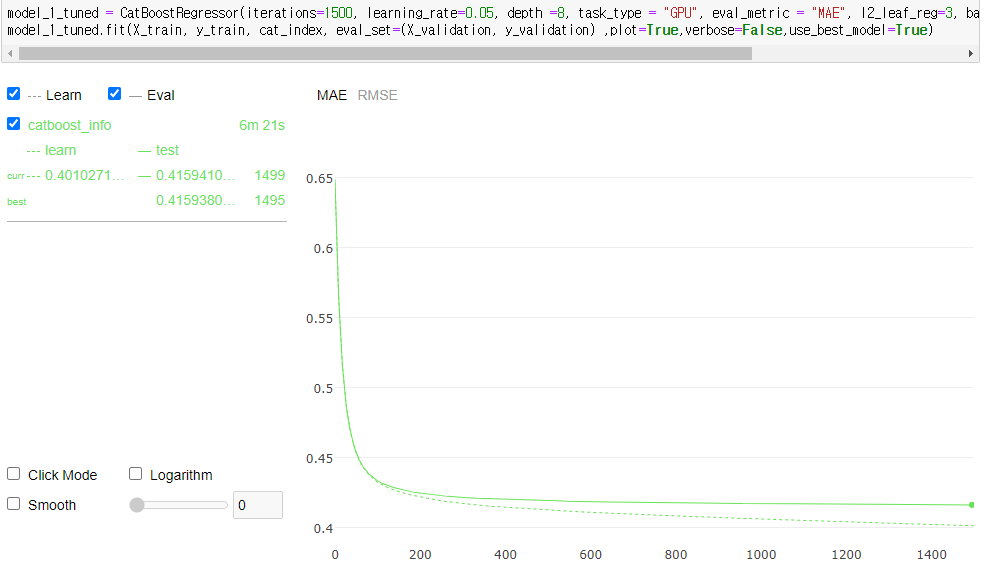
model_1_tuned.fit(X_train, y_train, cat_index, eval_set=(X_validation, y_validation) ,plot=True,verbose=False,use_best_model=True)
 <catboost.core.CatBoostRegressor at 0x151826f1cc8>
<catboost.core.CatBoostRegressor at 0x151826f1cc8>
확실히 더 좋은 성능을 보여준다.
print(model_1_tuned.evals_result_.keys())
print(model_1_tuned.learning_rate_,model_1_tuned.tree_count_)
mae_min_idx = np.argmin(model_1_tuned.evals_result_['validation']['MAE'])
print("Minimize MAE value is {:.6f} in Validation Set".format(model_1_tuned.evals_result_['validation']['MAE'][mae_min_idx]))
dict_keys(['learn', 'validation'])
0.05000000074505806 1496
Minimize MAE value is 0.415938 in Validation Set
df_test_['loss'] = np.exp(model_1_tuned.predict(df_test_.drop('loss', axis=1)))
df_test_.to_csv('kaggle_submission/submission_1_1.csv', sep = ',', columns = ['loss'], index='id')
kaggle collab 에서, 바로 적용시 활용하는 Code
!kaggle competitions submit -c allstate-claims-severity -f submission_1_1.csv -m “Catboost second submission (tuned)”
MAE 값이 basic PCA 했을때보다, param 수정한 버전이 좀 더 나은 결과를 보여준다.
skewed_cols를 제외한, 모델링
skewed_cols
['cont7', 'cont9']
df_train_ = traintest[traintest['isTrain']==True].drop(skewed_cols, axis = 1).set_index('id').drop('isTrain', axis = 1)
df_test_ = traintest[traintest['isTrain']==False].drop(skewed_cols, axis = 1).set_index('id').drop('isTrain', axis = 1)
cat_index = [i for i in range(0,len(df_train_.columns)-1) if 'cat' in df_train_.columns[i]]
cont_index = [i for i in range(0,len(df_train_.columns)-1) if 'cont' in df_train_.columns[i]]
X = df_train_.drop('loss', axis = 1)
y = np.log(df_train['loss'])
from sklearn.model_selection import train_test_split
X_train, X_validation, y_train, y_validation = train_test_split(X, y, train_size=0.8)
model_2 = CatBoostRegressor(iterations=600, learning_rate=0.05, depth =8, task_type = "GPU", eval_metric = "MAE", l2_leaf_reg=3, bagging_temperature=1,one_hot_max_size=0,use_best_model=True)
model_2.fit(X_train, y_train, cat_index, eval_set=(X_validation, y_validation),verbose=False ,plot=True)
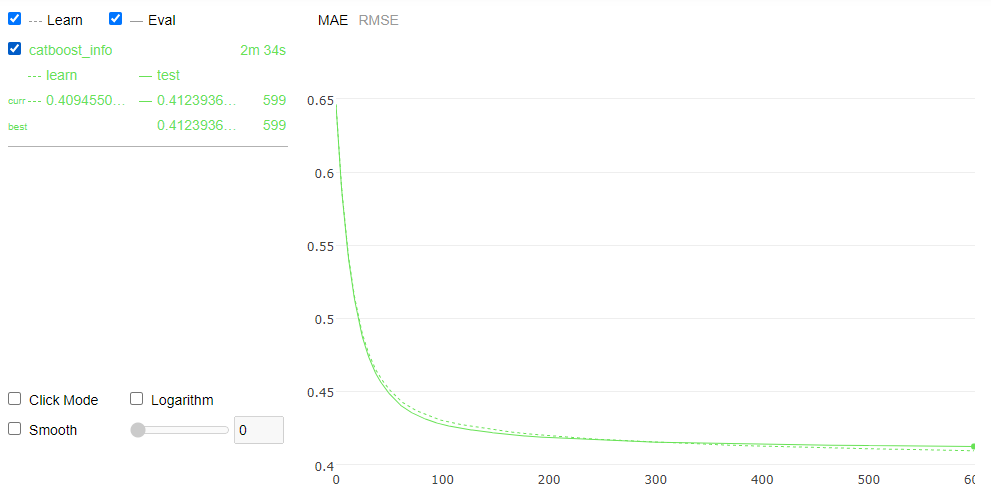 MetricVisualizer(layout=Layout(align_self=’stretch’, height=’500px’))
MetricVisualizer(layout=Layout(align_self=’stretch’, height=’500px’))
print(model_2.evals_result_.keys())
print(model_2.learning_rate_,model_2.tree_count_)
mae_min_idx = np.argmin(model_2.evals_result_['validation']['MAE'])
print("Minimize MAE value is {:.6f} in Validation Set".format(model_2.evals_result_['validation']['MAE'][mae_min_idx]))
dict_keys(['learn', 'validation'])
0.05000000074505806 600
Minimize MAE value is 0.412394 in Validation Set
df_test_['loss'] = np.exp(model_2.predict(df_test_.drop('loss', axis=1)))
df_test_.to_csv('kaggle_submission/allstate_submission_2.csv', sep = ',', columns = ['loss'], index=True)
3. Combine categorical variables
- train_enc : all categoried columns transformed to factorize_colum
train_enc.head(2)
| cat1 | cat2 | cat3 | cat4 | cat5 | cat6 | cat7 | cat8 | cat9 | cat10 | ... | cat107 | cat108 | cat109 | cat110 | cat111 | cat112 | cat113 | cat114 | cat115 | cat116 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ... | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | ... | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 1 |
2 rows × 116 columns
cum_cat_1 = []
for i in range(12,23):
cum_cat_1.append(‘cat’+str(i))
cum_cat_2 = []
for i in range(24,50):
cum_cat_2.append(‘cat’+str(i))
cum_cat_3 = []
for i in range(51,72):
cum_cat_3.append(‘cat’+str(i))
#create function - new traintest and new encoding
traintest = pd.concat([df_train, df_test], axis = 0)
cum_cat_1
['cat12',
'cat13',
'cat14',
'cat15',
'cat16',
'cat17',
'cat18',
'cat19',
'cat20',
'cat21',
'cat22']
## origin_code
cum_df_1 = train_enc[cum_cat_1].diff(axis=1).dropna(axis=1)
cum_df_1.columns = [col+'_cum' for col in cum_df_1.columns]
traintest = pd.concat([traintest, cum_df_1], axis=1).drop(cum_cat_1[1:], axis=1)
traintest[traintest['isTrain']==True].isnull().sum().sum()==0
dataframe 의 diff 함수는 row, columns 방향에 따라서, 전의 값과의 ‘차’ 를 return 한다.
dataframe.diff()
무슨 의도로, 이렇게 접근하는지 알 수 없기 때문에 종료한다.
Catboost tunning
from catboost import Pool as pool
import hyperopt
from hyperopt import hp, fmin, tpe, STATUS_OK, Trials
D_train = pool(X_train, y_train, cat_features = cat_index)
D_val = pool(X_validation, y_validation, cat_features = cat_index)
# number of random sampled hyperparameters
N_HYPEROPT_PROBES = 15
# the sampling aplgorithm
HYPEROPT_ALGO = tpe.suggest
def get_catboost_params(space):
params = dict()
params['learning_rate'] = space['learning_rate']
params['depth'] = int(space['depth'])
params['l2_leaf_reg'] = space['l2_leaf_reg']
params['one_hot_max_size'] = space['one_hot_max_size']
return params
# ---------------------------------------------------------------------
obj_call_count = 0
cur_best_loss = np.inf
log_writer = open( 'catboost-hyperopt-log.txt', 'w' )
def objective(space):
global obj_call_count, cur_best_loss
obj_call_count += 1
print('\nCatBoost objective call #{} cur_best_loss={:7.5f}'.format(obj_call_count,cur_best_loss) )
params = get_catboost_params(space)
# sorted_params = sorted(space.iteritems(), key=lambda z: z[0])
# params_str = str.join(' ', ['{}={}'.format(k, v) for k, v in sorted_params])
# print('Params: {}'.format(params_str) )
model = CatBoostRegressor(iterations=2000,
learning_rate=params['learning_rate'],
depth =int(params['depth']),
task_type = "GPU",
eval_metric = "MAE",
l2_leaf_reg=params['l2_leaf_reg'],
bagging_temperature=1,
one_hot_max_size=params['one_hot_max_size'],
use_best_model=True)
model.fit(D_train, eval_set=D_val, silent=True)
#y_pred = model.predict(df_test_.drop('loss', axis=1))
val_loss = model.best_score_['validation_0']['MAE']
if val_loss<cur_best_loss:
cur_best_loss = val_loss
return{'loss':cur_best_loss, 'status': STATUS_OK }
# --------------------------------------------------------------------------------
space ={
'depth': hp.quniform("depth", 4, 12, 1),
'learning_rate': hp.loguniform('learning_rate', -3.0, -0.7),
'l2_leaf_reg': hp.uniform('l2_leaf_reg', 1, 10),
'one_hot_max_size': hp.quniform("one_hot_max_size", 0, 15, 1)
}
trials = Trials()
best = hyperopt.fmin(fn=objective,
space=space,
algo=HYPEROPT_ALGO,
max_evals=N_HYPEROPT_PROBES,
trials=trials)
print('-'*50)
print('The best params:')
print( best )
print('\n\n')
hyperropt 라이브러리르 활용하는 방법이나, 현재 Local PC 의 한계로, Collab에서 수행하는 것을 권한다.
여기까지로 포스팅을 마친다.

Comments