Titanic Anlysis using Stacking & compare accuracy with using normal sckit-learn
Reference
- 허민석님의 Blog (https://github.com/minsuk-heo/kaggle-titanic/tree/master)
- 허민석님의 Youtube (https://www.youtube.com/watch?v=aqp_9HV58Ls)
- 머신러닝 스태킹 앙상블 (https://lsjsj92.tistory.com/559?category=853217)
Stacking 사용전, 기본 전처리 부분은 위 내용을 참조하였습니다.
import pandas as pd
train = pd.read_csv('../dataset/titanic_train.csv')
test = pd.read_csv('../dataset/titanic_test.csv')
import matplotlib.pyplot as plt
%matplotlib inline
import seaborn as sns
sns.set() # setting seaborn default for plots
train.isnull().sum()
PassengerId 0
Survived 0
Pclass 0
Name 0
Sex 0
Age 177
SibSp 0
Parch 0
Ticket 0
Fare 0
Cabin 687
Embarked 2
dtype: int64
test.isnull().sum()
PassengerId 0
Pclass 0
Name 0
Sex 0
Age 86
SibSp 0
Parch 0
Ticket 0
Fare 1
Cabin 327
Embarked 0
dtype: int64
posting 목적상, EDA 부분은 생략합니다. 원본을 참조하시면, 훨씬 훌륭하게 잘 정리되어 있습니다.
1. Feature 추가
train_test_data = [train, test] # combining train and test dataset
for dataset in train_test_data:
dataset['Title'] = dataset['Name'].str.extract(' ([A-Za-z]+)\.', expand=False)
train['Title'].value_counts()
Mr 517
Miss 182
Mrs 125
Master 40
Dr 7
Rev 6
Col 2
Mlle 2
Major 2
Sir 1
Countess 1
Capt 1
Lady 1
Don 1
Mme 1
Ms 1
Jonkheer 1
Name: Title, dtype: int64
Title map
Mr : 0
Miss : 1
Mrs: 2
Others: 3
categorical 하게 컬럼값을 처리한다.
title_mapping = {"Mr": 0, "Miss": 1, "Mrs": 2,
"Master": 3, "Dr": 3, "Rev": 3, "Col": 3, "Major": 3, "Mlle": 3,"Countess": 3,
"Ms": 3, "Lady": 3, "Jonkheer": 3, "Don": 3, "Dona" : 3, "Mme": 3,"Capt": 3,"Sir": 3 }
for dataset in train_test_data:
dataset['Title'] = dataset['Title'].map(title_mapping)
# delete unnecessary feature from dataset
train.drop('Name', axis=1, inplace=True)
test.drop('Name', axis=1, inplace=True)
sex_mapping = {"male": 0, "female": 1}
for dataset in train_test_data:
dataset['Sex'] = dataset['Sex'].map(sex_mapping)
2.Null 처리
# fill missing age with median age for each title (Mr, Mrs, Miss, Others)
train["Age"].fillna(train.groupby("Title")["Age"].transform("median"), inplace=True)
test["Age"].fillna(test.groupby("Title")["Age"].transform("median"), inplace=True)
# train.head(30)
print(train.groupby("Title")["Age"].agg("median"))
train.groupby("Title")["Age"].transform("median")[0:10] ## provide median value associated with Title-Age per each line
Title
0 30.0
1 21.0
2 35.0
3 9.0
Name: Age, dtype: float64
0 30.0
1 35.0
2 21.0
3 35.0
4 30.0
5 30.0
6 30.0
7 9.0
8 35.0
9 35.0
Name: Age, dtype: float64
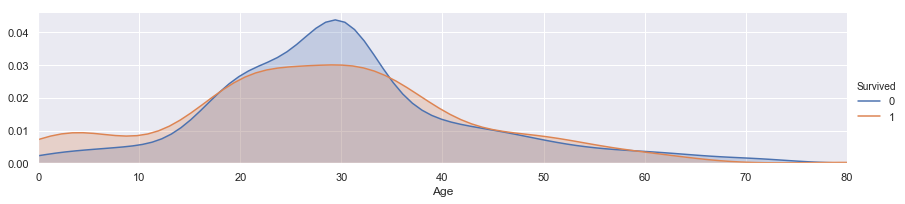
facet = sns.FacetGrid(train, hue="Survived",aspect=4)
facet.map(sns.kdeplot,'Age',shade= True)
facet.set(xlim=(0, train['Age'].max()))
facet.add_legend()
<seaborn.axisgrid.FacetGrid at 0x1d4a3025f60>
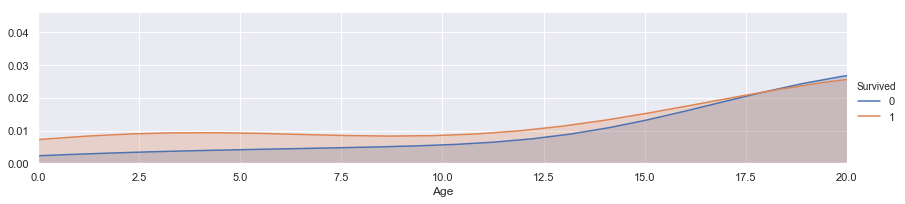
facet = sns.FacetGrid(train, hue="Survived",aspect=4)
facet.map(sns.kdeplot,'Age',shade= True)
facet.set(xlim=(0, train['Age'].max()))
facet.add_legend()
plt.xlim(0,20) ## This is an enlarged picture of the above.
(0, 20)
Pclass1 = train[train['Pclass']==1]['Embarked'].value_counts()
Pclass2 = train[train['Pclass']==2]['Embarked'].value_counts()
Pclass3 = train[train['Pclass']==3]['Embarked'].value_counts()
df = pd.DataFrame([Pclass1, Pclass2, Pclass3])
df.index = ['1st class','2nd class', '3rd class']
df.plot(kind='bar',stacked=True, figsize=(10,5))
<matplotlib.axes._subplots.AxesSubplot at 0x1d4a514e748>
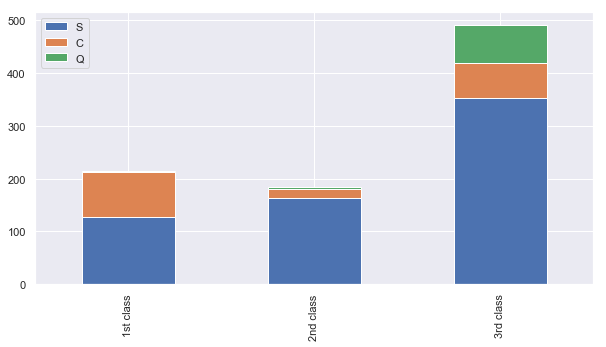
more than 50% of 1st class are from S embark
more than 50% of 2nd class are from S embark
more than 50% of 3rd class are from S embark
fill out missing embark with S embark
for dataset in train_test_data:
dataset['Embarked'] = dataset['Embarked'].fillna('S')
train.head()
| PassengerId | Survived | Pclass | Sex | Age | SibSp | Parch | Ticket | Fare | Cabin | Embarked | Title | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 1 | 0 | 3 | 0 | 22.0 | 1 | 0 | A/5 21171 | 7.2500 | NaN | S | 0 |
| 1 | 2 | 1 | 1 | 1 | 38.0 | 1 | 0 | PC 17599 | 71.2833 | C85 | C | 2 |
| 2 | 3 | 1 | 3 | 1 | 26.0 | 0 | 0 | STON/O2. 3101282 | 7.9250 | NaN | S | 1 |
| 3 | 4 | 1 | 1 | 1 | 35.0 | 1 | 0 | 113803 | 53.1000 | C123 | S | 2 |
| 4 | 5 | 0 | 3 | 0 | 35.0 | 0 | 0 | 373450 | 8.0500 | NaN | S | 0 |
embarked_mapping = {"S": 0, "C": 1, "Q": 2}
for dataset in train_test_data:
dataset['Embarked'] = dataset['Embarked'].map(embarked_mapping)
3. Feature engineering
for dataset in train_test_data:
dataset.loc[ dataset['Age'] <= 16, 'Age'] = 0,
dataset.loc[(dataset['Age'] > 16) & (dataset['Age'] <= 26), 'Age'] = 1,
dataset.loc[(dataset['Age'] > 26) & (dataset['Age'] <= 36), 'Age'] = 2,
dataset.loc[(dataset['Age'] > 36) & (dataset['Age'] <= 62), 'Age'] = 3,
dataset.loc[ dataset['Age'] > 62, 'Age'] = 4
# fill missing Fare with median fare for each Pclass
train["Fare"].fillna(train.groupby("Pclass")["Fare"].transform("median"), inplace=True)
test["Fare"].fillna(test.groupby("Pclass")["Fare"].transform("median"), inplace=True)
train.head(5)
| PassengerId | Survived | Pclass | Sex | Age | SibSp | Parch | Ticket | Fare | Cabin | Embarked | Title | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 1 | 0 | 3 | 0 | 1.0 | 1 | 0 | A/5 21171 | 7.2500 | NaN | 0 | 0 |
| 1 | 2 | 1 | 1 | 1 | 3.0 | 1 | 0 | PC 17599 | 71.2833 | C85 | 1 | 2 |
| 2 | 3 | 1 | 3 | 1 | 1.0 | 0 | 0 | STON/O2. 3101282 | 7.9250 | NaN | 0 | 1 |
| 3 | 4 | 1 | 1 | 1 | 2.0 | 1 | 0 | 113803 | 53.1000 | C123 | 0 | 2 |
| 4 | 5 | 0 | 3 | 0 | 2.0 | 0 | 0 | 373450 | 8.0500 | NaN | 0 | 0 |
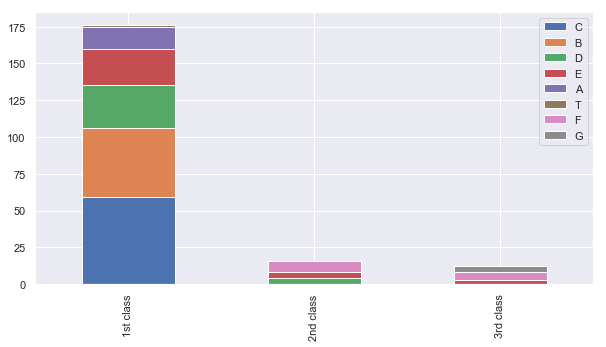
for dataset in train_test_data:
dataset['Cabin'] = dataset['Cabin'].str[:1]
Pclass1 = train[train['Pclass']==1]['Cabin'].value_counts()
Pclass2 = train[train['Pclass']==2]['Cabin'].value_counts()
Pclass3 = train[train['Pclass']==3]['Cabin'].value_counts()
df = pd.DataFrame([Pclass1, Pclass2, Pclass3])
df.index = ['1st class','2nd class', '3rd class']
df.plot(kind='bar',stacked=True, figsize=(10,5))
<matplotlib.axes._subplots.AxesSubplot at 0x1d4a50e97f0>
cabin_mapping = {"A": 0, "B": 0.4, "C": 0.8, "D": 1.2, "E": 1.6, "F": 2, "G": 2.4, "T": 2.8}
for dataset in train_test_data:
dataset['Cabin'] = dataset['Cabin'].map(cabin_mapping)
# fill missing Fare with median fare for each Pclass
train["Cabin"].fillna(train.groupby("Pclass")["Cabin"].transform("median"), inplace=True)
test["Cabin"].fillna(test.groupby("Pclass")["Cabin"].transform("median"), inplace=True)
train["FamilySize"] = train["SibSp"] + train["Parch"] + 1
test["FamilySize"] = test["SibSp"] + test["Parch"] + 1
facet = sns.FacetGrid(train, hue="Survived",aspect=4)
facet.map(sns.kdeplot,'FamilySize',shade= True)
facet.set(xlim=(0, train['FamilySize'].max()))
facet.add_legend()
plt.xlim(0)
(0, 11.0)
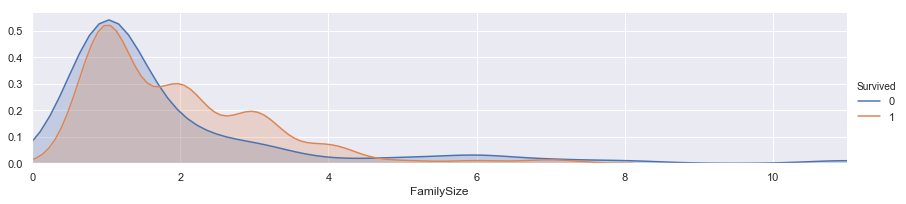
family_mapping = {1: 0, 2: 0.4, 3: 0.8, 4: 1.2, 5: 1.6, 6: 2, 7: 2.4, 8: 2.8, 9: 3.2, 10: 3.6, 11: 4}
for dataset in train_test_data:
dataset['FamilySize'] = dataset['FamilySize'].map(family_mapping)
train.head()
| PassengerId | Survived | Pclass | Sex | Age | SibSp | Parch | Ticket | Fare | Cabin | Embarked | Title | FamilySize | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 1 | 0 | 3 | 0 | 1.0 | 1 | 0 | A/5 21171 | 7.2500 | 2.0 | 0 | 0 | 0.4 |
| 1 | 2 | 1 | 1 | 1 | 3.0 | 1 | 0 | PC 17599 | 71.2833 | 0.8 | 1 | 2 | 0.4 |
| 2 | 3 | 1 | 3 | 1 | 1.0 | 0 | 0 | STON/O2. 3101282 | 7.9250 | 2.0 | 0 | 1 | 0.0 |
| 3 | 4 | 1 | 1 | 1 | 2.0 | 1 | 0 | 113803 | 53.1000 | 0.8 | 0 | 2 | 0.4 |
| 4 | 5 | 0 | 3 | 0 | 2.0 | 0 | 0 | 373450 | 8.0500 | 2.0 | 0 | 0 | 0.0 |
features_drop = ['Ticket', 'SibSp', 'Parch']
train = train.drop(features_drop, axis=1)
test = test.drop(features_drop, axis=1)
train = train.drop(['PassengerId'], axis=1)
Find Outlier && Remvoe Them
Using IQR : Inter Quntile Range (https://lsjsj92.tistory.com/556?category=853217)
백분위 수 : 데이터를 백등분 한 것
사분위 수 : 데이터를 4등분 한 것
중위수 : 데이터의 정 가운데 순위에 해당하는 값.(관측치의 절반은 크거나 같고 나머지 절반은 작거나 같다.)
제 3사분위 수 (Q3) : 중앙값 기준으로 상위 50% 중의 중앙값, 전체 데이터 중 상위 25%에 해당하는 값
제 1사분위 수 (Q1) : 중앙값 기준으로 하위 50% 중의 중앙값, 전체 데이터 중 하위 25%에 해당하는 값
사분위 범위 수(IQR) : 데이터의 중간 50% (Q3 - Q1)
- Top 25%:Q3 ~ Top 75%:Q1 so, IQR = Q3-Q1
- Limit max value = Q3 + IQR*1.5 (usually using 1.5)
- Limit min value = Q1 - IQR*1.5

## train 에서, Fare 만이 numeric 이라 볼 수 있다. FamilySize 는 가공변수고, 이미 order categori value 화 되었다. 다른 것도 마찬가지
# train.Fare
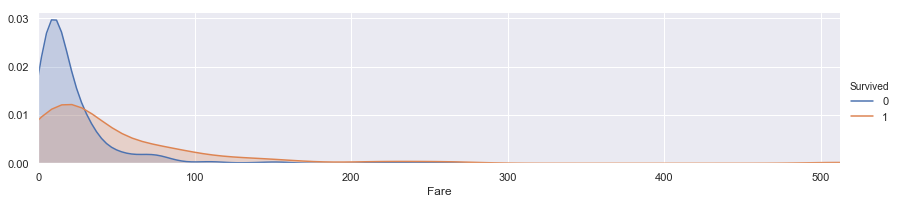
facet = sns.FacetGrid(train, hue="Survived",aspect=4)
facet.map(sns.kdeplot,'Fare',shade= True)
facet.set(xlim=(0, train['Fare'].max()))
facet.add_legend()
# plt.xlim(0)
<seaborn.axisgrid.FacetGrid at 0x1d4a6311e80>
facet = sns.FacetGrid(train, hue="Survived",aspect=4)
facet.map(sns.kdeplot,'Fare',shade= True)
facet.set(xlim=(0, train['Fare'].max()))
facet.add_legend()
plt.xlim(0,60)
(0, 60)
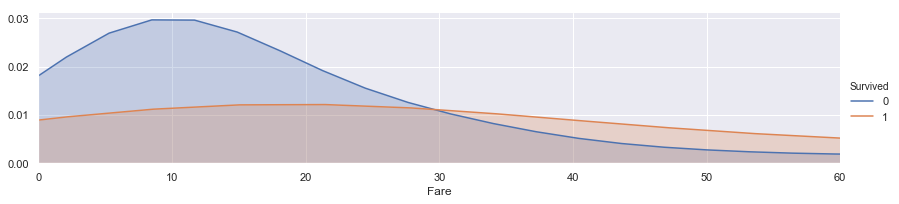
Fare feature looks having correlation with Surviced. so Check the outlier!!
train.Fare.describe().apply(lambda x : "{:.4f}".format(x))
count 891.0000
mean 32.2042
std 49.6934
min 0.0000
25% 7.9104
50% 14.4542
75% 31.0000
max 512.3292
Name: Fare, dtype: object
def ourlier_index(df,column,p):
q3 = df[column].quantile(0.75)
q1 = df[column].quantile(0.25)
iqr = q3 - q1
max_limit_val = q3+(iqr*p)
min_limit_val = 0 if q1-(iqr*p) < 0 else q1-(iqr*p)
a = (min_limit_val,max_limit_val)
print("min_limit_val {}".format(a[0]),"\t","max_limit_val {}".format(a[1]))
ix = df.loc[df.Fare < a[0]].index | df.loc[df.Fare > a[1]].index
left_ix = set(df.index)-set(ix)
return left_ix
train.groupby(by='Survived').Fare.describe().stack().apply(lambda x : "{:.4f}".format(x))
Survived
0 count 549.0000
mean 22.1179
std 31.3882
min 0.0000
25% 7.8542
50% 10.5000
75% 26.0000
max 263.0000
1 count 342.0000
mean 48.3954
std 66.5970
min 0.0000
25% 12.4750
50% 26.0000
75% 57.0000
max 512.3292
dtype: object
survived = 1,0 에 따라 아웃라이어가 다를 수 있으니, 다르게 접근하여 아웃라이어를 처리한다.
## when you want to find outlier and remove outlier value, run this code~
left_survive_ix = ourlier_index(train[train.loc[:,"Survived"]==1],"Fare",2.5)
left_die_ix = ourlier_index(train[train.loc[:,"Survived"]==0],"Fare",2.5)
print(len(left_survive_ix),len(left_die_ix))
left_survive_ix.intersection(left_die_ix) ## no dup index
train_01 = pd.concat([train.iloc[list(left_survive_ix)],train.iloc[list(left_die_ix)]],axis=0)
train_01.reset_index(drop=True,inplace=True)
print(train_01.shape)
train_01.head()
## when you don't want to find outlier and remove outlier value, run this code~
train_01 = train.copy()
train_data = train_01.drop('Survived', axis=1)
target = train_01['Survived']
train_data.shape, target.shape
((891, 8), (891,))
print(train_data.shape)
train_data.head(5)
(891, 8)
| Pclass | Sex | Age | Fare | Cabin | Embarked | Title | FamilySize | |
|---|---|---|---|---|---|---|---|---|
| 0 | 3 | 0 | 1.0 | 7.2500 | 2.0 | 0 | 0 | 0.4 |
| 1 | 1 | 1 | 3.0 | 71.2833 | 0.8 | 1 | 2 | 0.4 |
| 2 | 3 | 1 | 1.0 | 7.9250 | 2.0 | 0 | 1 | 0.0 |
| 3 | 1 | 1 | 2.0 | 53.1000 | 0.8 | 0 | 2 | 0.4 |
| 4 | 3 | 0 | 2.0 | 8.0500 | 2.0 | 0 | 0 | 0.0 |
total_y_sum = target.value_counts().sort_values(axis=0,ascending=False).sum()
target.value_counts().sort_values(axis=0,ascending=False)/total_y_sum
0 0.616162
1 0.383838
Name: Survived, dtype: float64
train_data = train_data.astype('float64')
Scaling
from sklearn.preprocessing import StandardScaler,MinMaxScaler
scaler = StandardScaler()
scaler.fit(train_data)
x_scaled = scaler.transform(train_data)
Modeling
# Importing Classifier Modules
from sklearn.neighbors import KNeighborsClassifier
from sklearn.tree import DecisionTreeClassifier
from sklearn.ensemble import RandomForestClassifier
from sklearn.naive_bayes import GaussianNB
from sklearn.svm import SVC
import numpy as np
Modeling - Cross Validation (K-fold)
from sklearn.model_selection import KFold
from sklearn.model_selection import cross_val_score
k_fold = KFold(n_splits=10, shuffle=True, random_state=0)
Modeling - kNN
clf = KNeighborsClassifier(n_neighbors = 13)
scoring = 'accuracy'
score = cross_val_score(clf, x_scaled, target, cv=k_fold, n_jobs=1, scoring=scoring)
print(score)
[0.83333333 0.7752809 0.80898876 0.82022472 0.83146067 0.83146067
0.76404494 0.84269663 0.82022472 0.82022472]
# kNN Score
round(np.mean(score)*100, 2)
81.48
Modeling - Decision Tree
clf = DecisionTreeClassifier()
scoring = 'accuracy'
score = cross_val_score(clf, x_scaled, target, cv=k_fold, n_jobs=1, scoring=scoring)
print(score)
[0.81111111 0.78651685 0.83146067 0.7752809 0.79775281 0.80898876
0.80898876 0.7752809 0.7752809 0.80898876]
# decision tree Score
round(np.mean(score)*100, 2)
79.8
Modeling - Ramdom Forest
clf = RandomForestClassifier(n_estimators=13)
scoring = 'accuracy'
score = cross_val_score(clf, x_scaled, target, cv=k_fold, n_jobs=1, scoring=scoring)
print(score)
[0.76666667 0.83146067 0.84269663 0.76404494 0.83146067 0.78651685
0.83146067 0.79775281 0.75280899 0.82022472]
# Random Forest Score
round(np.mean(score)*100, 2)
80.25
Modeling - Naive Bayes
clf = GaussianNB()
scoring = 'accuracy'
score = cross_val_score(clf, x_scaled, target, cv=k_fold, n_jobs=1, scoring=scoring)
print(score)
[0.82222222 0.74157303 0.7752809 0.75280899 0.74157303 0.83146067
0.80898876 0.82022472 0.82022472 0.83146067]
# Naive Bayes Score
round(np.mean(score)*100, 2)
79.46
Modeling - SVM
clf = SVC(gamma='auto')
scoring = 'accuracy'
score = cross_val_score(clf, x_scaled, target, cv=k_fold, n_jobs=1, scoring=scoring)
print(score)
[0.82222222 0.82022472 0.78651685 0.82022472 0.84269663 0.83146067
0.83146067 0.82022472 0.79775281 0.80898876]
round(np.mean(score)*100,2)
81.82
Stacking-Ensemble using K-Fold
하기 코드 이해를 위해, 몇가지 사전 연습
test_fold = np.random.randint(0,100,size=(10,3))
print(test_fold.shape)
(10, 3)
test_fold
array([[38, 79, 89],
[ 2, 66, 25],
[95, 56, 95],
[75, 2, 87],
[10, 34, 63],
[56, 46, 78],
[33, 38, 15],
[14, 82, 24],
[10, 63, 85],
[20, 23, 59]])
kk = KFold(n_splits=5, shuffle=False, random_state=42)
print("kk.get_n_splits :",kk.get_n_splits(test_fold))
for cnt , (train_idx,valid_idx) in enumerate(kk.split(test_fold)):
print("{} 번째 fold ".format(cnt),"폴더내, train 인덱스 {}".format(train_idx),"폴더내, train 인덱스 {}".format(valid_idx))
kk.get_n_splits : 5
0 번째 fold 폴더내, train 인덱스 [2 3 4 5 6 7 8 9] 폴더내, train 인덱스 [0 1]
1 번째 fold 폴더내, train 인덱스 [0 1 4 5 6 7 8 9] 폴더내, train 인덱스 [2 3]
2 번째 fold 폴더내, train 인덱스 [0 1 2 3 6 7 8 9] 폴더내, train 인덱스 [4 5]
3 번째 fold 폴더내, train 인덱스 [0 1 2 3 4 5 8 9] 폴더내, train 인덱스 [6 7]
4 번째 fold 폴더내, train 인덱스 [0 1 2 3 4 5 6 7] 폴더내, train 인덱스 [8 9]
여러개의 모델을 학습하고, 최종모델을 만들때, K-Fold에 기반하여 학습셋을 여러번 조직한다. 이때 편리하게 활용하기 위하여 별도 함수를 만들어 get_stacking_data() 관리한다
def get_stacking_data(model,x_train,y_train,x_test,n_folds=5):
kfold = KFold(n_splits=n_folds, random_state=42)
train_fold_predict = np.zeros((x_train.shape[0],1)) ## 폴드 횟수별, 검증 셋의 결과를 한줄로 담기 위한 shape
test_predict = np.zeros((x_test.shape[0],n_folds))
print("model : {}".format(model.__class__.__name__))
for cnt , (train_idx,valid_idx) in enumerate(kfold.split(x_train)):
x_train_ = x_train[train_idx]
y_train_ = y_train[train_idx]
x_val = x_train[valid_idx]
##학습
model.fit(x_train_,y_train_)
#해당 폴드에서 학습된 모델에다가 검증데이터로 예측 후 저장 이는 나중에 합쳐져서(쌓여서) new_x_train 이 된다.
train_fold_predict[valid_idx,:] = model.predict(x_val).reshape(-1,1)
#해당 폴드에서 생성된 모델에게 원본 테스트 데이터(x_test)를 이용해서 예측하고 저장
test_predict[:,cnt] = model.predict(x_test)
#for문 이후, test_predict 값은 평균을 내어서, 하나로 합친다.
test_predict_mean = np.mean(test_predict,axis=1).reshape(-1,1)
return train_fold_predict,test_predict_mean
clf_knn = KNeighborsClassifier(n_neighbors = 13)
clf_dt = DecisionTreeClassifier()
clf_rf = RandomForestClassifier(n_estimators=13)
clf_nb = GaussianNB()
clf_svm = SVC(gamma='auto')
staking_knn_train,staking_knn_test = get_stacking_data(clf_knn,x_scaled,target,xtest_scaled,n_folds=10)
staking_dt_train,staking_dt_test = get_stacking_data(clf_dt,x_scaled,target,xtest_scaled,n_folds=10)
staking_rf_train,staking_rf_test = get_stacking_data(clf_rf,x_scaled,target,xtest_scaled,n_folds=10)
staking_nb_train,staking_nb_test = get_stacking_data(clf_nb,x_scaled,target,xtest_scaled,n_folds=10)
staking_svm_train,staking_svm_test = get_stacking_data(clf_svm,x_scaled,target,xtest_scaled,n_folds=10)
model : KNeighborsClassifier
model : DecisionTreeClassifier
model : RandomForestClassifier
model : GaussianNB
model : SVC
print(staking_knn_train.shape,staking_knn_test.shape)
print(staking_dt_train.shape,staking_dt_test.shape)
print(staking_rf_train.shape,staking_rf_test.shape)
print(staking_nb_train.shape,staking_nb_test.shape)
print(staking_svm_train.shape,staking_svm_test.shape)
(891, 1) (418, 1)
(891, 1) (418, 1)
(891, 1) (418, 1)
(891, 1) (418, 1)
(891, 1) (418, 1)
new_x_train = np.concatenate([staking_knn_train,staking_dt_train,staking_rf_train,staking_nb_train,staking_svm_train],axis=1)
new_x_test = np.concatenate([staking_knn_test,staking_dt_test,staking_rf_test,staking_nb_test,staking_svm_test],axis=1)
print("origin x_train.shape {}".format(x_scaled.shape))
print("for stacking shape new_x_train.shape {}".format(new_x_train.shape)) ## total count of each clf is 5
print("for stackgin shape new_x_test.shape {}".format(new_x_test.shape))
origin x_train.shape (891, 8)
for stacking shape new_x_train.shape (891, 5)
for stackgin shape new_x_test.shape (418, 5)
shape 모양으로 stacking 의 구조를 잘 이해해야 한다.
stacking 모델은 개별 모델의 결과값을 feature 로 삼는데, k-fold 로 staking 방법을 사용하게 되면,
- 개별 분류기의 k-flod 검증 값들에 대한 예측값이 stacking train set = new_train 셋의 feature 1개가 된다.
여기선, 폴드가 10 이었기 때문에, (x_train[0]/10) 10조각으로 나누어져 validation 결과값이 만들어졌고, 이를 합치면, 모든 train 데이터의 validation 결과값 (x_train[0]) 나온다.
- 이런 값들이 모여서 stacking의 new_trainset이 되고, 여기선 개별분류기가 5개이니 최종 5개의 feature로 된 트렌인 셋이 만들어진다. -> (891,5)
- staking 의 new_x_test 는, 1개의 개별 분류기에서 10조각으로 나누어진 검증셋별로의 학습결과모델로 x_test 를 예측한 이후 이를 평균으로 만들고,
이를 합쳐서, 5개의 feature를 가진 new_x_test 가 된다. -> (418,5)
Testing
가장 잘 나온 SVM 모델로 구한값 – didn’t use stacking
clf = SVC(gamma='auto')
clf.fit(train_data, target)
test_data = test.drop("PassengerId", axis=1).copy()
test_data = test_data.astype('float64')
xtest_scaled = scaler.transform(test_data)
prediction = clf.predict(xtest_scaled) ## kaggle 제출결과 0.65071 outlier 처리 하기전이 더 나은듯. 과적합 된것으로 보인다.
참고로, 상기 원본 blog 자료에서, 추가로 야심차게 outlier 제거하고, scaling 변환 처리를 추가하여 나온 값이….원본보다, kaggle 결과가 더 나쁘다.
- 원본 blog : 0.78947
- outlier + scaling 처리 : 0.65071
- outlier (IQR변환 더크게) + scaling 처리 : 0.62679
lightgbm 모델로 구한값 – didn’t use stacking
from lightgbm import LGBMClassifier,plot_importance
lgb = LGBMClassifier(n_estimators = 400)
clf_lgb = lgb
clf_lgb.fit(train_data, target)
scoring = 'accuracy'
score = cross_val_score(clf_lgb, x_scaled, target, cv=k_fold, n_jobs=1, scoring=scoring)
print(score)
print(round(np.mean(score)*100,2))
[0.78888889 0.84269663 0.80898876 0.76404494 0.87640449 0.82022472
0.79775281 0.82022472 0.76404494 0.82022472]
81.03
test_data = test.drop("PassengerId", axis=1).copy()
test_data = test_data.astype('float64')
xtest_scaled = scaler.transform(test_data)
prediction = clf_lgb.predict(xtest_scaled) ## kaggle 제출결과 0.78468
submission = pd.DataFrame({
"PassengerId": test["PassengerId"],
"Survived": prediction
})
submission.to_csv('../dataset/submission.csv', index=False)
stacking 결과로 구하기
stacking_lgb = LGBMClassifier(n_estimators = 400)
stacking_lgb.fit(new_x_train,target)
LGBMClassifier(boosting_type='gbdt', class_weight=None, colsample_bytree=1.0,
importance_type='split', learning_rate=0.1, max_depth=-1,
min_child_samples=20, min_child_weight=0.001, min_split_gain=0.0,
n_estimators=400, n_jobs=-1, num_leaves=31, objective=None,
random_state=None, reg_alpha=0.0, reg_lambda=0.0, silent=True,
subsample=1.0, subsample_for_bin=200000, subsample_freq=0)
prediction = stacking_lgb.predict(new_x_test)
submission = pd.DataFrame({
"PassengerId": test["PassengerId"],
"Survived": prediction
})
submission.to_csv('../dataset/submission.csv', index=False) ## 0.79425 달성!!
# submission = pd.read_csv('submission.csv')
# submission.head()

Comments